Introduction
Studies have predicted that the cocoa beans market will increase by 7.3% annually until 2025 (Voora et al., 2019). Cocoa beans grow in tropical regions of Africa, Asia, Oceania, and Latin America (Kozicka et al., 2018). Worldwide, about 95% of cocoa is grown by smallholder farmers with an average size of 2–5 hectares of cultivated land (Escobar et al., 2020). In Colombia, this number reaches about 65,000 farmers, contributing to 95% of cocoa production (Abbott et al., 2017; Finagro, 2020), making Colombia the fifth highest producer of fine flavor cocoa worldwide and the third in Latin America (Ortiz et al., 2014). Considering these facts, the Colombian Ministry of Agriculture and Rural Development has passed public policies such as the Cocoa development ten year plan 2012–2021 (MinAgricultura, 2008). Nevertheless, in Colombia, there is a need for a standardized cocoa cultivation process to increase the quality of cocoa beans (Colombia productiva, 2019).
The quality of cocoa and its derived products is dependent on various factors, from the bean harvest to the end of the industrial process, fermentation being perhaps the most important factor. The fermentation process of cocoa is primarily spontaneous due to the artisanal and traditional practices of farmers (Melo-Pereira et al., 2013; Schwan et al., 2015; Pereira et al., 2016). Consequently, various indigenous microorganisms are involved in the spontaneous fermentation of cocoa beans (yeasts; lactic acid bacteria (LAB), acetic acid bacteria (AAB), and, possibly, species of bacillus and other bacteria; and filamentous fungi) (Mendoza et al., 2022). Fermentation begins with the growth of yeasts that produce ethanol under anaerobiosis conditions from sugars present in the pulp (mucilage). Subsequently, LAB converts ethanol and citric acid into lactic acid and other substances, and AAB produces acetic acid via the oxidation of ethanol, leading to a noticeable increase in temperature (Chagas et al., 2021). Conventional fermentation techniques of cocoa beans carried out by some small-holders do not always result in good-quality fermented cocoa beans (low repeatability between cocoa fermentation batches, risk of contamination, disagreeable tastes, and odors) (Díaz-Muñoz and De Vuyst, 2022). Using starter cultures could lead to standardized products with good quality and sensory properties (Lima et al., 2011). Thus, knowledge about microbiota and its contribution to cocoa beans fermentation is crucial (Leal et al., 2008; Lefeber et al., 2012; Crafack et al., 2013; Saltini et al., 2013). Some authors have reported that Saccharomyces, Hanseniaspora (anamorph Kloeckera), Pichia membranifaciens, Pichia kudriavzevii, and some Candida spp. are predominant yeasts found in the early stages of cocoa beans fermentation (Ho et al., 2014; Neto et al., 2017; Mendoza et al., 2022). However, the ecology and population dynamics of specific microbiota in the Florida region (Valle del Cauca, Colombia) have not yet been studied.
This study was carried out to understand how microorganisms and their proliferation influence cocoa beans fermentation in Florida (Colombia). This work aimed to investigate controlled fermentation of cocoa beans with selected yeasts as starter cultures via integrating microbiological, biochemical, and chromatographic analyses.
Material and methods
Plant material for yeast isolation
During the primary cocoa harvest of 2018 (April and June), three spontaneous fermentation processes of cocoa beans were carried out in three small production units of cocoa farmers: sampling point no. 1 (M1): 3.270716 3·16′14.58 ″, −76.21647676·12′59.31″; sampling point no. 2 (M2): 3.264080 3·15′50.69″, −76.21104076·12′39.74″; and sampling point no. 3 (M3): 3.262311 3·15′44.32″, −76.21364876··12′49.13″ in Florida (Valle del Cauca, Colombia). The spontaneous fermentation processes were carried out for 5 days and performed according to the study conducted by Garcia-Gonzalez et al. (2021). At the physiological maturation stage, cocoa (Theobroma cacao L.) cobs of Trinitario clones were collected and broken using handmade cutting tools. In the production units M1, M2, and M3, 12 kg (in a 20 l cylindrical high-density polyethylene container), 10 kg (wooden box of 1 m3), and 20 kg (in a 200 l cylindrical high-density polyethylene container) of cocoa beans were fermented, respectively, according to the artisanal cocoa beans fermentation practices of the farmers.
Cocoa fermentation under laboratory conditions with selected yeasts
The steps involved in the yeast starter culture test were of the following order: 1) counting, isolation, purification, and biochemical identification of yeasts, 2) selection of ethanol-producing yeasts, 3) selection of thermotolerant yeasts, and 4) evaluation of the selected yeasts in controlled fermentation of cocoa.
Microorganism counting, isolation, and purification
For the isolation and counting of microorganisms, 10 g of cocoa beans was used. Microorganisms were separated by extracting the pulp from nibs (cotyledon), and then, the pulp was macerated and homogenized in 90 ml (10−1 dilution) of 0.1% peptone water using a stirrer at 140 rpm for 30 min (Merck, EEUU). Subsequently, serial dilutions (v/v) of the pulp were made from 10−1 to 10−6. Isolation and enumeration of yeasts were performed using Sabouraud Chloramphenicol Agar (Sharlau, Spain). Yeasts were incubated at 30°C for 48 h, and then, the number of colony-forming units (CFU) per gram of fresh cocoa was determined. To identify the isolated yeasts, biochemical tests (API 20C AUX gallery – Bio-Merieux, Brazil) were performed according to the manufacturer’s instructions. Each kit contained 32 cupules, which were inoculated with 135 μl of yeast suspensions prepared in the basal medium and incubated at 30°C. Reactions were observed after 24 and 48 h. Each receptacle was identified by entering the 10-digit numerical profile in the API web software. Mesophilic aerobes were counted in Tryptone Agar yeast extract (Himedia, India) and incubated at 30°C for 48 h. After incubation, the number of CFU per gram of fresh cocoa was recorded. The counting of microorganisms was performed in duplicate.
Selection of ethanol-producing yeasts
Six presumptive Saccharomyces cerevisiae yeast strains were identified using the biochemical tests; these strains were used in ethanol production. The following simulated cocoa medium was used (Adler et al., 2013): 27.5 g/l of glucose (Oxoid, Germany), 25 g/l of fructose (Merck, Germany), 10 g/l citric acid (Sigma-Aldrich, Germany), 5 g/l yeast extract (Merck, Germany), 5 g/l soy peptone (Neogen, USA), 0.5 g/l magnesium sulfate heptahydrate (Panreac, Spain), 0.2 g/l of manganese sulfate monohydrate (Panreac, Spain), and 1 ml/l of Tween 80 (Panreac, Spain). Using a 2 N NaOH solution, the pH of the medium was adjusted to 3.8. The fermentations were carried out for 24 h, and five samples were taken at different time points (0, 2, 4, 8, and 24 h). Cell concentration of the samples was evaluated by counting in a Neu Bauer chamber, and optical density (OD540) was determined using a spectrophotometer (Mapada, China). The simulated cocoa fermentations were carried out in triplicate.
After 24-h fermentation, ethanol production was measured for each yeast strain. The percentage of ethanol was determined using the specific gravity method (Muchtaridi et al., 2013). In brief, the fermented samples were centrifuged at 4000 rpm for 10 min, and 40 ml of the supernatant was diluted at a 1 : 4 ratio of supernatant and distilled water. Of the diluted solution, 100 ml was distilled to obtain 100 ml of the distilled solution (J.P. Selecta, Spain). Then, the percentage of ethanol was measured in the distilled solution using a densimeter (Metler Toledo 3PX, Japan). Ethanol measurements were performed in duplicate.
Selection of thermotolerant aromatic yeasts
Five yeasts biochemically identified as Kloeckera ssp. were evaluated for their thermotolerance (Meersman et al., 2015). These yeasts were screened for temperature tolerance in assay tubes. Strains were pregrown in the simulated cocoa medium (Adler et al., 2013) for 24 h at 30°C and then inoculated at an initial OD at 550 nm (OD550) of 0.1 in the simulated cocoa medium. The yeasts were subsequently incubated at 30, 36, and 42°C for 24, 48, and 72 h, and cell growth was quantified using OD at 550 nm (OD550). The tests were performed in duplicate. The difference between growth at time (t) and the initial time (to) was determined (ΔOD550) as follows:
Cocoa fermentation under controlled conditions with selected yeasts
Under controlled fermentation conditions, starter cultures were added at specified concentrations. S. cerevisiae CLV09 and Kloeckera ssp. CLV20 were selected. The former was selected for its ability to produce ethanol (Koff et al., 2017; Kouamé et al., 2021), and the latter was selected for its heat resistance at 42°C (Meersman et al., 2015). Both these strains have been reported as predominant yeasts during cocoa beans fermentation. In addition, both strains contribute to the fermented fruity and floral aroma of cocoa beans (Nara et al., 2015). Three serial dilutions were made for the counting of the cultured yeasts (10−1, 10−2, and 10−3). The last dilution was made with methylene blue (Chemi, India) (0.01%) in a ratio of 1 : 1.
Fermentations were performed in 12 × 12 × 12 cm cubic Sajo wooden boxes (capacity 1728 cm3) constructed by the authors of this study. These boxes were loaded with 500 g of cocoa beans. Fermentations were monitored for 5 days. Two fermentations were carried out: starter culture fermentation (F1) and spontaneous fermentation (F2-control). F1 fermentation was inoculated with Kloeckera ssp. CLV20 and S. cerevisiae CLV09 strains. The initial concentration was adjusted to 106 CFU/g of cocoa beans, and fermentations were carried out in duplicate. The incubator set point was increased 5°C above ambient temperature to a final temperature of 55°C. The yeasts were inoculated in the fermentation boxes, and samples were taken daily to determine pH, titratable acidity (Steinau, 2017), and concentration of reducing sugars (Miller, 1959), yeast and mesophilic aerobes.
Physicochemical tests
The pH of fresh cocoa beans was determined (Steinau, 2017). Five cocoa beans were taken, and the pulp was separated from the nibs. Then, the pulp was macerated separately and diluted in 50 ml of water. Subsequently, the pH was measured in duplicate using a BANTE 210 potentiometer (China).
Reducing sugars were determined using the dinitrosalicylic acid method (Miller, 1959). To 10 g of previously macerated cocoa, 90 ml of distilled water was added, and the solution was stirred for 30 min. Subsequently, successive dilutions were made up to 10−3, and the reducing sugar concentration was determined.
Organic acid concentration determination
Fermented cocoa beans were stored at −20°C (at each fermentation time). Four cocoa beans were weighed, and their pulp/nibs proportion was determined (the pulp was separated from the nibs, and each part was weighed). Samples of 1.00 ± 0.01 g of cocoa pulp and cocoa nibs were macerated separately with 5 ml of type 1 Ultrapure Water for 2 min (Smart2Pure 3 UV/UF – Thermo Scientific, Spain). Subsequently, mortar washes were conducted with 15 ml of type 1 Ultrapure Water. Each fraction was mixed in a vortex for 5 min and then subjected to an ultrasound bath for another 5 min. The homogenate was vacuum-filtered on a qualitative No. 2 filter (Advantec, Japan). In addition, the samples of cocoa nibs were filtered through SPE C18 Cartridges (Macherey Nagel, Germany), and then, solutions of cocoa pulp and cocoa nibs were filtered back through a 0.25 μm membrane (Sartorius Biolab, Germany). Finally, ultra-high-performance liquid chromatography (uHPLC Thermo Scientific, Spain) analysis was carried out.
Lactic acid, acetic acid, and ethanol concentrations were determined using chromatographic analysis (uHPLC) in a Dionex Ultimate 3000 chromatograph with a refractive index detector at 35°C (ERC Refractomax 520). Chromatographic separations were achieved using the Carbohydrate H + HyperRez XP cation exchange column (300 × 7.7 mm), with a particle size of 8 μm (Thermo Scientific, USA). Lactic acid, acetic acid, and ethanol were eluted with a 0.5 mM sulfuric acid mobile phase at a flow rate of 0.6 ml/min at 65°C. Analytes were identified according to the respective standard retention time, and the concentration of the analytes was determined by the extrapolation of peak areas with the calibration curve. Individual samples were analyzed in duplicate, and average values were reported.
Statistical analysis
The following variables were measured during fermentation (5 days): reducing sugars (mg/g), pH in cocoa pulp and cocoa nibs, yeasts, and concentration of mesophilic aerobes (CFU/g). All measurements were performed in duplicate. Mixed models with random coefficients were used to analyze these variables. Independent samples ANOVA was established for this group of variables. R statistical software, version 3.5.0, was used for the analysis. The “nlme” library was used to adjust mixed models of random coefficients, and the “stat” library was used with the Kruskal function. In nonparametric ANOVA tests (Team, 2013), 5% significance was used for the hypothesis contrast.
Results and discussion
Biochemical identification of yeasts
Thirty-two Saccharomyces spp., Kloeckera spp., Candida spp., and Rhodotorula spp. were isolated from spontaneous fermentations carried out in three small production units of cocoa farmers (Florida, Vale del Cauca, Colombia) and biochemically identified (Table 1), of which 50% were Candida ssp., 9.4% were Rhodotorula ssp., 18.8% were Saccharomyces ssp., and 18.8% were Kloeckera ssp. Candida spp. were observed in concentrations of 102–106 CFU/g of cocoa beans in the three sampling points (M1, M2, and M3). In M2 and M3, the yeasts were present from the beginning of fermentation up to 96 h, whereas in M1, the yeasts were present at 48 and 96 h. Rhodotorula spp. strains were identified in M2 at concentrations from 102 to 105 CFU/g of cocoa beans. Saccharomyces ssp. were observed at 96 h at 105 CFU/g of cocoa beans in M2 and at concentrations of 102–105 CFU/g at M3. Kloeckera ssp. were identified in M1, M2, and M3 at concentrations of 104–106 CFU/g of cocoa beans between 24 and 72 h. According to Papalexandratou and De Vuyst (2011), the most frequently isolated yeast species in cocoa beans fermentation are S. cerevisiae, Hanseniaspora guilliermondii (anamorph Kloeckera apis), Hanseniaspora opuntiae, Pichia fermentans, Pichia kluyveri, P. kudriavzevii, P. membranifaciens, and Wickerhamomyces anomalus. In a study of Fernández et al. (2016), Candida species are the most frequent in spontaneous fermentation (25 equipment and handling-related samples, and 115 environmental samples such as flowers, leaf, and cocoa pod surfaces). Delgado-Ospina et al. (2020) reported that the most frequent and dominant yeast species in cocoa beans fermentation of the Criollo variety are P. kudriavzevii (40%) and S. cerevisiae (28%) along with other yeasts such as Pichia manshurica (9%), Candida parapsilosis (7%), and W. anomalus (4%). The presence of Hanseniaspora spp. in cocoa beans fermentation has also been documented (Lagunes Gálvez et al., 2007; Lefeber et al., 2012; De Melo et al., 2013).
Table 1
Biochemical identification of yeasts and isolation conditions
Selection of ethanol-producing yeasts
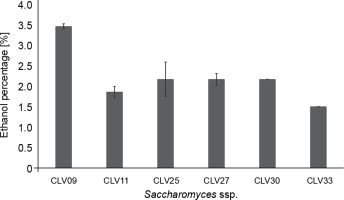
In total, 32 strains were isolated, of which six S. cerevisiae yeasts were selected based on biochemical identification (Table 1). Saccharomyces spp. were selected due to their predominant role in fermentation and higher ability to produce ethanol compared to other species (Visintin et al., 2017; Nara et al., 2015). Previous studies have reported ethanol production in cocoa beans fermentation by adding S. cerevisiae (Lefeber et al., 2012; Díaz-Muñoz and De Vuyst, 2022). The biochemical test revealed that the yeasts identified as S. cerevisiae were CLV09, CLV11, CLV25, CLV27, CLV30, and CLV33 (Table 1). Among these strains, Saccharomyces spp. CLV09 showed the highest ethanol production in the simulated cocoa medium (3.5% v/v) (Fig. 1). Saccharomyces spp. CLV25, CLV27, and CLV30 showed identical ethanol production (2.2% v/v) after 24 h of fermentation, whereas strains CLV11 and CLV33 showed the lowest ethanol production (1.8 and 1.5% v/v, respectively). Koffi et al. (2018) assessed the ethanol production potential of 113 yeast strains isolated from cocoa fermentation, of which 19 strains of S. cerevisiae showed yields above 4.8%.
Selection of thermotolerant yeasts
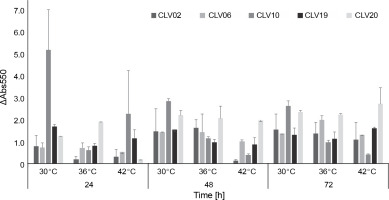
Hanseniaspora spp., detected during cocoa fermentation, has been reported as a significant yeast species associated with S. cerevisiae (Batista et al., 2019; Moreira et al., 2013; Ramos et al., 2014). Yeasts produce esters and higher alcohols, which contribute to the mixture of volatile aroma compounds of chocolate aroma (Crafack et al., 2013). The aromatic qualities of Hanseniaspora spp. are well known (Gamero et al., 2016; Rossouw and Bauer, 2016; Ooi et al., 2020). In cocoa bean fermentation, using microorganisms with aromatic potential is an important factor. However, some strains do not withstand high fermentation temperatures (Papalexandratou and Nielsen, 2016). Hence, yeasts with both thermotolerance and high aroma production were selected. Thermotolerance is an important factor because the temperature could increase up to 45°C during the fermentation period (Horta-Téllez et al., 2019). Thus, in the present study, thermotolerance of Hanseniaspora spp. at different temperatures was evaluated. In all the Hanseniaspora spp. strains evaluated (CLV02, CLV06, CLV10, CLV19, and CLV20), growth was observed in the temperature range assessed from 24 to 72 h (Fig. 2). CVL20 and CVL19 strains showed the highest thermotolerance at 42°C at 72 h of growth (ΔAbs550 = 2.74 and ΔAbs550 = 1.63, respectively), whereas CVL10 strain showed the lowest thermotolerance among the strains evaluated (ΔAbs550 = 0.43). These results are consistent with those of the study by Hamdouche et al. (2015), in which H. opuntiae was one of the predominant species in cocoa fermentation and survived bean drying. Hanseniaspora ssp. CLV20 was selected to evaluate fermentation with the starter yeast due to its high thermotolerance.
Cocoa fermentation under controlled conditions with selected yeasts
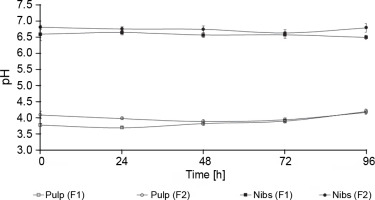
No statistically significant differences in the pH of the cocoa pulp were observed between F1 and F2 (P > > 0.05). However, statistically significant differences in the pH of the nibs were observed between F1 and F2, where F1 showed a lower pH (6.50 ± 0.05) compared with F2 (6.80 ± 0.07) (Fig. 3). Racine et al. (2019) reported that the initial pH of the simulated pulp media was approximately 3.6 and then increased to a maximum final mean pH value of 4.6 and that the mean pH value of the solution made from the nibs was 5.4 and declined throughout fermentation to a final mean pH value of 4.6. The cocoa bean pulp is permeable to acetic acid, which then passes into the nibs (cotyledon) after 3 days, killing the embryo and lowering the pH (Peláez et al., 2016). Laboratory-scale fermentation has limitations in contrast to conventional field fermentation. These limitations are chiefly related to the use of piles of boxes. Approximately 150 kg of fresh wet cocoa beans is used in the onfarm cocoa fermentation process. The fermentation can reach the appropriate temperature, pH, and moisture under the abovementioned conditions (Lee et al., 2018). The low quantity of cocoa pulp used in the present study may explain the limitation in the decrease in the pH of the nibs and the increase in the pH of the cocoa pulp.
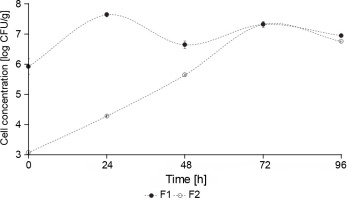
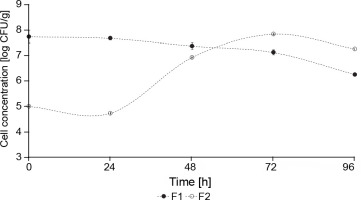
The concentration of yeasts in F1 fermentation (starter culture) was significantly higher than in F2 fermentation (control – spontaneous fermentation) (P < 0.05) (Fig. 4). At the beginning of F1 fermentation, the yeast concentration was 5.93 log CFU/g, whereas F2 fermentation showed an initial concentration of 3.07 CFU/g. During the 5 days of fermentation, F1 showed a constant growth between 7 and 6 log CFU/g, and F2 reached 7 log CFU/g after 72 h of fermentation. This behavior can be explained by the fact that the yeast inoculum (S. cerevisiae and Hanseniaspora spp.) was added at a concentration of 106 CFU/g in F1 fermentation, whereas the yeast concentration in F2 fermentation was lower since it was a spontaneous fermentation. Nara et al. (2015) reported a similar behavior where a higher concentration of S. cerevisiae, Hanseniaspora uvarum, and Pichia kluyveri were observed in the inoculated stage fermentation than in the control cocoa fermentation.
The concentration of mesophilic aerobes can be an indicator of other microorganisms that can develop under cocoa beans fermentation conditions (Lagunes Gálvez et al., 2007). The concentration of mesophilic aerobes was nearly constant during F1 fermentation and ranged from 7.75 log CFU/g at the beginning of the fermentation to 6.26 log CFU/g at 96 h (Fig. 5). In F2 fermentation, the concentration of mesophilic aerobes was 7.84 log CFU/g in 72 h, starting from 5 log CFU/g. As shown in Fig. 4, a nearly constant yeast concentration was observed during the 96 h of fermentation (in F1 fermentation), whereas the concentration of mesophilic aerobes in F1 fermentation started to decrease after 48 h of fermentation (Fig. 5). This behavior can be explained by the dominance of S. cerevisiae over other microbial species during fermentation, which is attributable to its enhanced fermentative potential and ability to withstand the increasingly adverse conditions established in the medium as fermentation progresses (Nara et al., 2015).
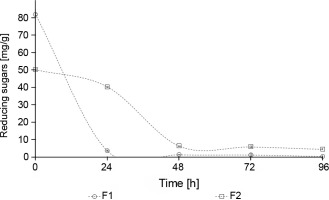
The concentration of reducing sugars in F1 fermentation was significantly lower than in F2 fermentation (P < 0.05) (Fig. 6). In the first 24 h of fermentation, 95.5% of reducing sugars were consumed in F1 fermentation, whereas in F2 fermentation, only 19.6% of sugars were consumed. These results are consistent with the pattern of growth observed for yeasts and mesophilic aerobes, which was accompanied by a deceleration of microbial growth from 48 to 72 h. These results are related to the decrease in sugar levels during the fermentation process from 24 to 48 h (Fig. 6).
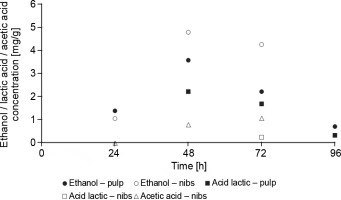
In F1 fermentation, metabolites produced during fermentation (ethanol, lactic acid, and acetic acid) were detected (Fig. 7), whereas these metabolites were not detected in F2 fermentation. The absence of these metabolites in F2 fermentation (spontaneous fermentation) can be attributable to the low concentrations of yeasts and mesophilic aerobes in the first 48 h of cocoa beans fermentation, during which a significant amount of ethanol is produced (Nara et al., 2015). The ethanol concentration in the cocoa pulp at 24, 48, and 72 h of F1 fermentation was 1.39, 3.58, and 2.21 mg/g, respectively. In the nibs, at 24, 48, and 72 h of F1 fermentation, it was 1.79, 4.79, and 4.26 mg/g, respectively. Nara et al. (2015) evaluated the behavior of S. cerevisiae, P. kluyveri, and H. uvarum during spontaneous and inoculated fermentation and reported similar values of ethanol production (6, 8, and 8 mg/g of the cocoa pulp; and 3, 7, and 5 mg/g of the nibs at 24, 48, and 72 h, respectively). In F1 fermentation, the ethanol concentration detected in the nibs was higher than in the pulp. The high ethanol and acetic acid concentrations in the first hours of cocoa beans fermentation in the nibs can lead to early death of the cocoa embryo. Likewise, lactic acid concentrations in the pulp were 2.21, 1.69, and 0.33 mg/g at 48, 72, and 96 h of fermentation, respectively. Lactic acid was not detected at 24 h of fermentation, which coincides with the low concentration of mesophilic aerobes. The lactic acid concentration in the nibs was 0.25 mg/g at 72 h of fermentation. Lefeber et al. (2012) reported similar results in a study in which starter microorganisms (S. cerevisiae H5S5K23, Lactobacillus fermentum 222, and Acetobacter pasteurianus 386B) were added as a starter culture in a box (500-kg scale) and constant lactic acid concentrations of 2.5 mg/g were observed during 48, 72, and 96 h of cocoa pulp fermentation. Lee et al. (2018) reported acetic acid concentrations of 2 and 3 mg/g at 48 and 72 h in cocoa beans fermentation, respectively. In the present study, acetic acid was detected at the concentrations of 0.8 and 1.08 mg/g at 48 and 72 h in F1 fermentation. At 96 h of F1 fermentation, ethanol was not detected in the fermented cocoa beans (nibs). This phenomenon can be explained by the oxidation of ethanol to acetic acid and the conversion of acetic acid and lactic acid to aromatic compounds as a result of pyruvate metabolism (De Vuyst and Weckx, 2016). Metabolites (acetic acid, lactic acid, ethanol, and others) formed in the pulp of cocoa beans diffuse into the nibs during fermentation, with an increase in temperature triggered by the oxidation of ethanol, inducing the death of the cocoa embryo (De Vuyst and Weckx, 2016), which – after fermentation – becomes the fermented cocoa bean (Mendoza et al., 2022).
Conclusions
Thirty-two Saccharomyces spp., Kloeckera spp., Candida spp., and Rhodotorula spp. were isolated from spontaneous fermentations of cocoa beans in production units of cocoa smallholders (Florida, Colombia). Saccharomyces spp. CVL09 showed the highest ethanol production (3.5%). Hanseniaspora ssp. CVL20 showed the highest thermotolerance at 42°C for 72 h. The starter culture fermentation process (F1) showed a higher production of organic acids than the spontaneous fermentation process (F2). Based on these results, Saccharomyces ssp. and Hanseniaspora ssp. strains can be used together as starter cultures in cocoa beans fermentation. Further studies on Saccharomyces spp. and Hanseniaspora spp. should be conducted under environmental conditions. Studies on yeast, LAB, and AAB strains (isolated and inoculated in cocoa beans fermentation) need to be carried out to evaluate the behavior of starter cultures with all the microorganisms involved in cocoa beans fermentation.