Introduction
The wide application of synthetic dyes for product colorization (Zeng et al., 2015) in industries such as textile, paper, plastics, printing, leather, cosmetics, and pharmaceuticals has led to the generation of waste-waters containing high loads of dye pollutants rich in aromatic compounds (Rezaei et al., 2015; Blánquez et al., 2019). The presence of these dyes in water, even at very low concentrations, is highly visible and therefore esthetically unpleasant (Jawad et al., 2016). In addition, the discharge of such wastewaters poses severe problems to the environment as some dyes and their biodegradation products are mutagenic and carcinogenic in nature (Imran et al., 2016).
Methylene blue is an organic dye known mostly for its extensive use in textile, leather, paper, plastics, and paint industries (Contreras et al., 2019). These industries use large amounts of methylene blue, with an estimate concentration of 10 to 200 mg/l being discharged into the environment, particularly in China and India (Habibi and Narimanirad, 2019; Sarkar et al., 2017). Methylene blue is highly recalcitrant and toxic in nature, and direct consumption of methylene blue-rich water may result in diarrhea, vomiting, nausea, inflammation of the stomach, hallucination, cyanosis, jaundice, tissue necrosis, methemoglobinemia, and excessive sweating (Bharti et al., 2019). Considering these adverse effects, methylene blue ingestion in rats, based on the safety data sheet, was established at the median lethal dose (LD50) value of 1.18 g/kg, which is consistent with European Commission Regulation (EU) 2015/830 (Lacasa et al., 2019). Therefore, in order to protect the interest of humans and the environment, effluents containing methylene blue need to be treated prior to discharge into the surface waters.
The complicated and stable structure of aromatic and heterocyclic methylene blue often makes the dye resistant to treatment (Holkar et al., 2016). Therefore, conventional physicochemical treatment methods such as electrocoagulation cannot effectively decolorize methylene blue. Electrocoagulation uses iron or aluminum as an anode, and the electrolytic oxidation process forms hydroxides. These hydroxides then react with the pollutants present in water through two mechanisms, namely surface complexation or electrostatic attraction. Ozonation is another physicochemical treatment method that can be used to decolorize methylene blue. In this method, ozone attacks the azoic –N=N– double bond that is linked with color and decolorizes the dye. On the other hand, the photochemical method uses H2O2 in the presence of Fe2+ (Fenton process) to generate hydroxy radicals (•OH), a highly oxidant species with high standard potential of 2.8 V. This facilitates the nonselective mineralization of almost all organic chemicals to CO2, H2O, and inorganic ions. (Al-Baldawi et al., 2018). This results in incomplete decolorization of methylene blue and the formation of toxic byproducts, which is environmentally unacceptable. Moreover, even though the physicochemical methods of wastewater treatment are easy to use, these methods may not always be cost-effective due to the consumption of expensive reagents, high capital and operating costs, and time-consuming removal methods (Bhatia et al., 2017).
Qualities such as high competency, simplicity, and low cost make adsorption as one of the most strategic methods for dye removal from wastewaters (Chandane and Singh, 2016; Beakou et al., 2017; Surip et al., 2020). Among the adsorbents, the abundance of natural adsorbents, namely agricultural wastes, is exploited for methylene blue removal because of their environmental friendliness, low cost, and sustainability (Daud et al., 2018; Stavrinou et al., 2018; Kadhom et al., 2020). The agricultural sector in Malaysia is thriving, especially in the area of fruit commodity production (Mansor et al., 2018; Baharin et al., 2020). Consequently, this results in voluminous fruit waste (0.310 million tonnes of sugarcane bagasse, 0.239 million tonnes of coconut waste, 803 938 tonnes of banana waste, and 275 732 tonnes of pineapple waste in 2015), thus presenting an opportunity for it to be used as an effective adsorbent (Mansor et al., 2018; Baharin et al., 2020). These wastes contain functional groups such as hydroxy, phenols and carboxyl groups, which enable pollutants to attach to the agricultural wastes (Daud et al., 2018). Biological methods may, however, present as a more appropriate approach for decolorization of methylene blue, wherein the colorant is converted into byproducts (Katheresan et al., 2018; Joshi et al., 2020). Biological materials such as algae, bacteria, fungi, and yeasts can be applied to degrade and absorb a variety of synthetic dyes such as methyl red, methyl orange, Acid red, Remazol blue, and Congo red (Bhatia et al., 2017; Sarkar et al., 2017). Biological methods of dye removal break bonds of the chromophore group, leading to the removal of color. The synthetic dyes are degraded to relatively less toxic inorganic compounds, depending on the type of micro-organism used and the process conditions (Bhatia et al., 2017). In the first step of the catabolism process, organisms such as algae, bacteria, fungi, and yeasts break the azo bonds in the dye structure to form amines. In the second step, under aerobic environment, aromatic amines are converted to small nontoxic molecules (Bhatia et al., 2017).
Biological methods of dye removal by using bacteria, in particular, were found to be largely nonhazardous, depending on the bacterial strain used; for example Rhodococcus pyridinivorans strain UCC 0003 used in the present study is nonpathogenic. As shown previously, this method is an economically viable solution for decolorization and biodegradation of various dyes from industrial wastewater (Saratale et al., 2013). These procedures result in a lesser sludge production and require less energy and chemicals than that required for the physicochemical treatment methods (Martínez-Sánchez et al., 2018). Furthermore, bacteria are easy to cultivate and can propagate rapidly as compared to other microorganisms such as microalgae and fungi (Bhatia et al., 2017). This is because bacteria show fast and abundant growth under optimum temperature and oxygen conditions, the medium for bacterial growth is usually cheap and easily available, and many protocols are available for bacterial cultivation.
Many studies have reported the remarkable roles of bacteria such as Staphylococcus hominus, Vibrio sp., Proteus vulgaris, Dietzia sp., Pseudomonas aeruginosa, Bacillus subtilis, Stenotrophomonas sp., and Halomonas sp. in efficient decolorization of dyes, and there are many other bacterial species that have developed their enzyme systems for degrading dye compounds (Mansur et al., 2017; Roy et al., 2018; Karnwal, 2019; Mandic et al., 2019). Bacteria possess multiple mechanisms to remove dyes, and because they are easy to use, they have advantages in terms of commercial value (Sarim et al., 2019).
We have previously reported efficient decolorization of methylene blue by using R. pyridinivorans strain UCC 0003 cells immobilized in polyurethane foam; this is a robust bacterial strain capable of functioning as a biocatalyst for dye removal even though the strain was not isolated from dye-contaminated sources (Nallapan Maniyam et al., 2020a). Therefore, the first aim of the present study was to elucidate the mechanism of methylene blue decolorization by R. pyridinivorans strain UCC 0003 through UV-Vis spectrophotometric analysis. The second aim of the present study was to investigate the viability of using agricultural waste materials such as fruit wastes, which have little or no economic value, as an additional carbon source for the growth of dye-degrading bacteria and thus help to minimize the agricultural waste disposal problem (Barathi and Ramesh, 2013). This study also intended to determine the rate of decolorization of methylene blue through a kinetic study.
Agricultural wastes were primarily designed as adsorbents for the removal of methylene blue, as reported in previous studies (Miraboutalebi et al., 2017; Choi and Yu, 2018). A recent study by Lawagon and Amon (2019) revealed that magnetic rice husk ash could enhance the rate of adsorption of methylene blue by 10 to 200 mg/l. The adsorption of methylene blue occurred on the surface of the adsorbent. In most cases, the dye did not undergo chemical changes through the adsorption mechanism; specifically, no physical adsorption or chemical adsorption was observed. A research study on the adsorptive removal of methylene blue did not discuss the toxicity of the dye as this aspect can only be evaluated on the remaining aqueous solution of methylene blue or dye wastewaters. In addition, metabolites such as toxic amines could be formed after decolorization of methylene blue and sometimes accumulate during the biological removal of methylene blue using, for example, bacteria under anaerobic conditions; this could make the industrial scale-up of the process more challenging (Sarkar et al., 2017; Al-Tohamy et al., 2020). Therefore, the present study evaluated the toxicity of the solution resulting from the biodecolorization of methylene blue by using R. pyridinivorans strain UCC 0003. The final aim of the study was to perform a phytotoxicity analysis using Vigna radiata and Triticum aestivum seeds. These findings could serve as preliminary data to evaluate the feasibility of using biologically treated industrial wastewaters containing dyes for application in irrigation systems.
Materials and methods
Chemicals
Analytical grade chemicals were used without any further purification. These chemicals were commercially available and obtained either from Merck (Germany), Sigma (USA), or Fisher Scientific (Singapore). The chemicals included medium components (nutrient broth and minimal salt medium), methylene blue, and commercial carbon and nitrogen sources.
Microorganism
R. pyridinivorans strain UCC 0003 (Hasdianty et al., 2020) was isolated from a sludge sample from Kuala Lukut, Negeri Sembilan, Malaysia, and kindly provided by the Culture Collection Unit of Institute of Bio-IT Selangor, Universiti Selangor. This strain has been deposited in GenBank® (NCBI) under the gene accession number MN56068.
Decolorization of methylene blue by R. pyridinivorans strain UCC 0003 under different culture conditions
Agitation
A seed culture of R. pyridinivorans strain UCC 003 was prepared in a 250 ml conical flask. Next, 50 ml of 8 g/l nutrient broth medium (basic medium composed of 0.5% (w/v) simple peptone and 0.3% (w/v) beef extract) was prepared. R. pyridinivorans strain UCC 0003 was inoculated into the medium. The incubation was performed at 160 rpm and 30°C (S1600R (B3L) Lab Companion, Jeiotech, Korea) for 24 h until an optical density (OD) of 1 was reached, which was measured spectrophotometrically at 600 nm.
Seed culture (10% (v/v)) was placed aseptically (Bio-safety Cabinet 4ft-EN12469, ESCO Micro (M) Sdn. Bhd.) in 50 ml medium (in 250 ml conical flask) comprising 8 g/l nutrient broth. Stock solution of 2 g/l methylene blue was filter sterilized (0.2 micron, Sterlitech, USA) and added to the nutrient broth medium so that a final concentration of 0.1 g/l methylene blue was achieved. To evaluate the effect of agitation on the decolorization of methylene blue, the mixtures were incubated at 30°C for 24 h at 160 rpm. For static condition, the media were left unagitated under the same experimental conditions. The experiments were performed in triplicate.
Aerobic/anaerobic conditions
To examine the effects of anaerobic and aerobic culture conditions on the decolorization of methylene blue by R. pyridinivorans strain UCC 0003, the methods described by Guadie et al. (2017) were followed, and the reactions were performed in a 1 l stirred tank bioreactor (Biostat B-plus, Sartorius Stedim, Germany) without agitation. The bioreactor was equipped with temperature monitoring and dissolved oxygen control systems. A polarographic electrode, calibrated using saturated air (100% saturation) and water saturated with nitrogen (0% saturation), was used to determine the dissolved oxygen tension (DOT) level in the culture. This protocol was followed as the fluctuation in the concentration of dissolved oxygen could greatly affect the degree of decolorization of methylene blue. The anaerobic culture condition was maintained using pure nitrogen, bubbled until the dissolved oxygen reached zero. This procedure was not used for the anoxic culture. The continuous aeration condition (airflow rate of 3 l/min) was used to maintain the aerobic culture. All experiments were performed at 30°C and pH 7.0 under an initial methylene blue concentration of 0.1 g/l. The percentage of methylene blue removal and dissolved oxygen was monitored as a function of time.
At 0°C and 24 h, 2 ml samples were removed and centrifuged at 14 000 × g for 15 min at 4°C (Eppendorf 5702R, South Asia) to separate the cell mass from the resulting clear supernatant. The supernatant was subjected to a UV-Vis spectrophotometric analysis after dilution, if necessary. The absorbance was measured at 200–700 nm wavelength (Biomate 3 Thermo Fisher Scientific, USA; absorbance reading ranging from –3.99 to 3.99 absorbance unit with the least error between 0.1 and 0.8 absorbance units). The quantification of the abiotic loss of methylene blue was established using separate incubation in the absence of R. pyridinivorans strain UCC 0003 and the use of heat-killed microorganisms. The formula, as shown in Equation 1, was used to determine the decolorization activity in terms of percentage; the experiments were performed in triplicate.
Optimization of culture conditions for enhanced decolorization of methylene blue by R. pyridinivorans strain UCC 0003
The ability of R. pyridinivorans strain UCC 0003 to perform enhanced decolorization of methylene blue was evaluated by varying the effects of operational and environmental conditions. These batch experiments were conducted using the seed culture of R. pyridinivorans strain UCC 0003 cultivated in nutrient broth and incubated for 24 h, as reported earlier. The culture was resuscitated under static conditions in a minimal salt medium (MSM) containing 5 g/l KH2PO4, 5 g/l Na2HPO4, and 0.5 g/l methylene blue (except for the initial dye concentration parameter evaluation). Each experiment was repeated three times, and the results were averaged. The same experimental protocols were used in the control experiment, except for the addition of seed cultures. The time required for complete decolorization of methylene blue, measured at 660 nm wavelength, was recorded.
The following decolorization parameters were established to evaluate the decolorization performance of the locally isolated actinomycete.
The viability of recycling the local isolate as a biocatalyst was assessed in the present study. Under the optimum conditions (35°C, pH 6, 5% (v/v) banana peel extract, 16% (v/v) inoculum concentration, and no addition of nitrogen sources), the decolorization of 0.75 g/l methylene blue was observed. Samples were withdrawn at every 15-min interval until 0.75 g/l methylene blue was entirely decolorized. Following complete decolorization, fresh methylene blue aliquot (0.75 g/l) was added to the same medium, and the decolorization was recorded with samples taken at every 15-min interval until complete decolorization was achieved. This process was repeated for several cycles until the optimum percentage of decolorization decreased to below 60%.
Kinetic study
The kinetic study was performed by monitoring the rate of disappearance of methylene blue in a culture of R. pyridinivorans strain UCC 0003 over a certain period of incubation time in sealed vials at 30°C. The substrate in this study was methylene blue. This protocol was performed to establish the kinetic parameters of decolorization of methylene blue by the abovementioned bacterial strain by using varying concentrations of methylene blue as the substrate, from 0.4 to 10.0 g/l. The reaction mixture contained 8 g/l nutrient broth adjusted aseptically with 10% (v/v) seed culture of R. pyridinivorans strain UCC 0003. The experiment was performed in triplicate. Kinetics parameters, namely Km and Vmax, for methylene blue were calculated using nonlinear regression fitting of the Lineweaver-Burk transformation of the Michaelis-Menten equation using Prism 5 with at least eight initial substrate concentrations.
Phytotoxicity study
The phytotoxicity study was performed using V. radiata and T. aestivum by following the protocols established by Nallapan Maniyam et al. (2018).
Statistical analysis
All experiments were conducted in triplicate. Experimental errors were estimated and shown as error bars (standard error) typically with three determinations. All data were analyzed using SPSS version 17.0. Comparison between groups was performed using the Duncan analysis (values with different superscript letters are significantly different). A one-way ANOVA test (95% confidence interval) was used to evaluate the differences between groups, and P < 0.05 was considered to be statistically significant. Kinetic data were analyzed using GraphPad software version 5.02.
Results and discussion
Decolorization of methylene blue by R. pyridinivorans strain UCC 0003 under different culture conditions
The decolorization of methylene blue by R. pyridinivorans strain UCC 0003 was performed under anoxic (static), shaking, aerobic, and anaerobic culture conditions. This particular strain grew well under static conditions when methylene blue was used as an auxiliary source of carbon and nitrogen, resulting in a cell concentration of 0.75 ± 0.15 mg/ml dry cell weight after an incubation period of 24 h; this in turn supported the efficient decolorization of methylene blue, which reached 86 ± 0%. In addition, the methylene blue removal activity of 57 ± 0% was achieved after 24 h of incubation under anaerobic culture conditions.
It was interesting to note that the decolorization of methylene blue under a lower concentration of dissolved oxygen in anoxic (0.25 ± 0.0 mg/l) and anaerobic (0.01 ± 0.0) culture conditions contributed to a higher percentage of dye removal by this local isolate. From this finding, it was concluded that the efficiency of methylene blue decolorization increased due to the disruption of the chemical structure of the colorant by R. pyridinivorans strain UCC 0003 under the anoxic condition as compared to that under shaking and aerobic conditions. Similar findings were reported by Chakraborty et al. (2016), wherein complete decolorization of 0.05 g/l methylene blue was achieved by Escherichia coli and Bacillus subtilis under static conditions. In addition, another study by Fulekar et al. (2013) reported that it took 7 days to degrade 69% of 0.02 g/l methylene blue by a Pseudomonas sp. isolated from soil. Chen et al. (2015) indicated an almost complete removal of 0.025 g/l methylene blue (95%) by Bacillus thuringiensis 016 after an incubation period of 48 h. Another research study conducted by Rizqi and Purnomo (2017) showed that a brown-rot fungus Daedalea dickinsii could decolorize approximately 54% of 0.1 g/l methylene blue after 14 days. Therefore, the present study reports a more superior finding of methylene blue decolorization than that reported earlier.
In contrast, a significantly lower methylene blue removal activity (20%, P < 0.05) was observed when the decolorization was performed under shaking conditions as compared to that under static conditions. A substantial reduction in the methylene blue-decolorizing activity by 77% was observed even though a considerable cell concentration of 0.68 ± 5.15 mg/ml dry cell weight was obtained when R. pyridinivorans strain UCC 0003 was incubated at an agitation of 160 rpm for 24 h. Similarly, a methylene blue removal activity of 27% was observed under an aerobic culture condition with cell concentration of 0.77 ± 3.26 mg/ml dry cell weight. These findings clearly indicate that the presence of high concentrations of dissolved oxygen (1.81 ± 0.12 mg/l and 1.96 ± 0.16 mg/l) for shaking and aerobic culture conditions, respectively, impeded efficient decolorization of methylene blue by R. pyridinivorans strain UCC 0003. This may be due to the presence of oxygen (high redox-potential electron acceptor) under aerobic conditions, which may lead to the inhibition of the dye reduction mechanism (Das et al., 2015). Throughout the experimental period, the concentration of 0.1 g/l methylene blue in the control group without R. pyridinivorans strain UCC 0003 remained almost the same, and the heat-inactivated culture only showed a decolorization activity of 1%. These observations ruled out the abiotic loss of methylene blue. On the basis of the results, the decolorization of methylene blue by R. pyridinivorans strain UCC 0003 under anoxic culture conditions was adopted for further investigation of optimization.
Mechanism of methylene blue decolorization
The mechanism of methylene blue decolorization under static and shaking conditions was determined by spectrophotometric scanning in the range of 200 to 700 nm of culture supernatants withdrawn at 0 h and after 24 h incubation period, as shown in Figure 1. The methylene blue spectrum in the visible region obtained from the methylene blue sample before being subjected to decolorization by R. pyridinivorans strain UCC 0003 exhibited three major peaks with the maximum absorbance recorded at 220 nm, 340 nm, and 660 nm. The major peak at 660 nm disappeared after an incubation period of 24 h under static conditions, suggesting that the immediate elimination of the colorant was due to the active presence of the locally isolated R. pyridinivorans strain UCC 003 as the biocatalyst. After decolorization by R. pyridinivorans strain UCC 0003, the peak at 660 nm progressively shifted to a shorter wavelength (Fig. 1) because of the hypsochromic effect (Zuo et al., 2014). In contrast, the minimal decline in the strength of the peak at 660 nm, which is attributed to the heteropolyaromatic linkage (Zuo et al., 2014), was observed when the methylene blue solution was tested by the UV-Vis spectrum analysis after 24 h incubation under shaking condition. This finding indicated that the decolorization of methylene blue under the agitated mode was not favorable and resulted in an ineffective disruption of the heteropolyaromatic linkage, leading to the presence of the major peak, although of a slightly reduced absorbance intensity.
Fig. 1
UV–Vis spectra profiles of the parent dye and methylene blue decolorization metabolites under static and shaking conditions; incubation was performed at 30°C and pH 7 for 24 h with 0.1 g/l methylene blue; control was established in the absence of R. pyridinivorans strain UCC 0003; error bars represent standard error between three determinations
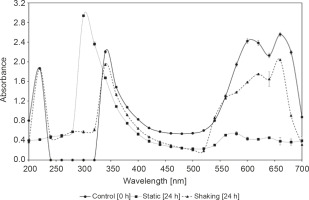
In addition, with the decrease in the absorption peak at the wavelength of 660 nm, the peak at 220 nm completely disappeared, while the peak at 340 nm was shifted to 300 nm and increased significantly when the methylene blue sample treated with R. pyridinivorans strain UCC 0003 under the static mode was subjected to a UV-Vis analysis (Chen et al., 2015). The peaks at 220 nm and 340 nm wavelengths, however, did not alter for the methylene blue sample obtained after biological treatment by this local isolate under agitated culture conditions; this indicated that this was not the preferred mode for the decolorization of methylene blue by R. pyridinivorans strain UCC 0003.
The existence of absorbance peak alteration coupled with a complete removal of the major visible light absorbance peak was possibly due to the absorbance of metabolites or degraded fragments of dye molecules in the decolorized solution of methylene blue in the static mode within 24 h of incubation period, as shown in Figure 1. This obvious evidence suggested that the occurrence of methylene blue depletion might be largely contributed by R. pyridinivorans strain UCC 0003 as the biocatalyst used to convert the colorant to less toxic products. Further confirmation and metabolite identification of methylene blue and its degradation products will be was carried out in the future using Fourier Transform Infrared (FTIR) Spectroscopy and mass spectroscopy analyses to support the premise that the methylene blue removal occurred through biodegradation by the locally isolated actinomycete.
Optimization of culture conditions for enhanced decolorization of methylene blue by R. pyridinivorans strain UCC 0003
Effect of temperature
To study the effect of temperature on the removal of the colorant, the decolorization of methylene blue by R. pyridinivorans strain UCC 0003 was evaluated at 20 to 45°C at the initial concentration of 0.5 g/l. The methylene blue removal increased from 11 ± 1 % to 70 ± 1% when the temperature was increased from 20 to 30°C, resulting in an increase of 536% in the methylene blue decolorizing activity, as shown in Table 1. Similar findings were reported by Bharti et al. (2019), who showed the occurrence of optimal decolorization of methylene blue of 68% at 30°C by using immobilized cells of Alcaligenes faecalis in biochar. A direct relationship was observed between the environmental temperature and microbial activity (Balraj et al., 2016). Adaptation through biochemical or enzymatic mechanisms occurs as the microbial cells actively respond to temperature changes. In the present study, R. pyridinivorans strain UCC 0003 could achieve an optimal metabolic function at 30°C, making it capable enough to secrete the required enzymes needed for the decolorization of methylene blue (Balraj et al., 2016). Moreover, this finding supported the typical characteristic of a mesophile, which was observed in R. pyridinivorans strain UCC 0003 isolated from a sludge sample (Maniyam et al., 2013; Nallapan Maniyam et al., 2015). A control group without the addition of methylene blue was used. The growth pattern of R. pyridinivorans strain UCC 003 was similar to that observed in the decolorization experiment, where the optimum growth was observed at 30°C.
Table 1
Effect of different culture conditions on the decolorization of methylene blue by R. pyridinivorans strain UCC 0003
[i] The control was established in the absence of R. pyridinivorans strain UCC 0003; a negligible loss of methylene blue was detected in the control system (less than 1% removal); the percentage of methylene blue removal is expressed as means of triplicate samples ± standard errors; statistically significant differences (P < 0.05) were observed among the tested groups; different letters indicate a significant difference in the tested factors; the factors were ranked from the most favored to the least favored following the alphabetical order in superscript format (a – most favored, least favored – e)
A further increase in the temperature beyond 30°C decreased the extent of methylene blue removal. When the temperature further increased to 45°C, the percentage of decolorization of methylene blue decreased to 12 ± 1%. As shown in earlier reports, loss of cell viability and inhibition of enzyme activity occur at higher temperatures and thus contribute to the decline in the percentage of methylene blue removal (Talha et al., 2018). In addition, reduced mobility and thermal breakdown of bacterial cells could be another factor that influences the decrease in the decolorization of methylene blue at elevated temperatures (Bharti et al. 2019). A similar finding was reported by Zeng et al. (2015) who documented reduced degradation of methylene blue by Phanerochaete chrysosporium to 60% at 45°C from 85% at 35°C by using 0.4 g/l methylene blue after an incubation period of 22 days.
Effect of pH
A wide pH value ranging from 4 to 10 was selected to study the effects of pH on the decolorization of methylene blue. The results indicated that R. pyridinivorans strain UCC 0003 was unable to decolorize methylene blue effectively under acidic conditions, achieving methylene blue-decolorizing activity of 13 ± 2% and 21 ± 1% at pH values of 4 and 5, respectively. This finding was not surprising as it is a distinct characteristic of an actinomycete to favorably grow in alkaline conditions (Nallapan Maniyam et al., 2013).
It was interesting to note that increasing the pH to a range between 6 to 8 reduced the time needed for enhanced decolorization of methylene blue by 50%, indicating that this pH range was the most suitable for the decolorization of methylene blue. The optimum methylene blue removal efficiency was recorded at pH 6, with methylene blue-decolorizing activity of 74 ± 1% after an incubation period of 12 h as shown in Table 1. A change in the incubation time from 12 to 24 h at pH 6, 7, and 8 yielded a similar methylene blue-decolorizing activity of 80% after 24 h. When the decolorization of methylene blue by R. pyridinivorans strain UCC 0003 was performed at pH 9 and 10, 40 ± 0% and 41 ± 1% removal of methylene blue was noted, respectively, after 24 h of incubation period; this indicated a high decolorizing activity under alkaline conditions. The use of natrium hydroxide in large amounts in the scouring, washing, and mercerizing steps of textile operation (Prasad and Aikat, 2014) makes most textile wastewaters highly alkaline in nature. Therefore, the ability of R. pyridinivorans strain UCC 003 to decolorize methylene blue under alkaline conditions is advantageous for practical applications in treating textile wastewater.
There is a possibility that methylene blue may be discolored due to changes in pH values. Hence, control experiments were conducted to exclude the likelihood of the occurrence of this scenario and to further confirm whether methylene blue decolorization was indeed due to microbial activity. These controls were established to evaluate the effect of pH on the visible absorption of methylene blue when assayed between pH 4 and 10 in a 20 mM sodium dimethylsuccinate buffer. As expected, no shift in pH values was recorded when the pH was measured after methylene blue removal by R. pyridinivorans strain UCC 0003 at pH 4 to 10, indicating that the local isolate was alone responsible for the decolorization of the colorant (Saratale et al., 2013).
Effect of carbon and nitrogen sources
The management of industrial residues favors economic and environmental sustainability of processes; therefore, there is a worldwide trend to develop more efficient production chains (Almeida et al., 2018). Considering this aspect, it is appropriate to use agro-industrial wastes that are huge sources of readily available carbon and may be used as substrates or supporting agents for the decolorization of methylene blue by R. pyridinivorans strain UCC 0003. The application of agroindustrial wastes as carbon sources in enzyme production processes reduces the cost of production and helps to solve problems associated with environmental effects on industrial production (Unuofin et al., 2019).
In recent years, there has been an increasing trend in the production of fruits in Malaysia, and this has inevitably generated a voluminous amount of fruit waste (Mansor et al., 2018). Therefore, the continuous availability of fruit waste should be smartly exploited as an energy source (Mansor et al., 2018), thereby converting waste to wealth. In the present study, the effects of supplementing 5% (v/v) agricultural wastes such as banana peel, coconut husks, sugarcane bagasse, and orange peel on the decolorization of methylene blue by the locally isolated R. pyridinivorans strain UCC 0003 were evaluated. The use of all types of carbon source (conventional and agricultural wastes) supported the growth of R. pyridinivorans strain UCC 0003 with no significant difference (P > 0.05), resulting in an optical density (measured at 600 nm) of 0.215–0.219 as compared to the control system with an optical density of 0.087 after 2 h of incubation period. This result indicated that the local strain could use agricultural wastes as growth nutrients, which in turn facilitated the decolorization of methylene blue and therefore provided a cheaper alternative for the cultivation of R. pyridinivorans strain UCC 0003. It was interesting to note that a methylene blue-decolorizing activity of above 80% was observed when banana peel was used as a carbon source, with a significantly (P < 0.05) higher percentage of methylene blue decolorization (100%) after 2 h of incubation period (Table 1). In addition, the supplementation of banana peel as a carbon source managed to significantly reduce the time taken for complete decolorization of methylene blue from 12 to 2 h.
Efficient decolorization of methylene blue was also observed in the presence of orange peel, coconut husks, and sugarcane bagasse, resulting in methylene blue removal of 81 ± 2%, 91 ± 2%, and 84 ± 3%, respectively, after 2 h of incubation. These findings indicated that the use of easily available and cheaper nutrient sources such as banana peel, orange peel, coconut husks, and sugarcane bagasse readily supported the decolorization of methylene blue by R. pyridinivorans strain UCC 0003. In addition, this research may provide an alternative to the use of agro residues as an additional carbon source before their disposal, thus promoting an environmentally friendly wastewater treatment process (Saratale et al., 2013). Maltose, glucose, and sucrose did not yield satisfactory results when supplemented as carbon sources, showing less than 30% methylene blue removal after 2 h of incubation period. Similar findings were reported by Unuofin et al. (2019), indicating that the use of grape stalks enhanced the production of laccase by 3.22-fold in comparison to a simple carbon source. In addition, a greater activity and a robust laccase production of 38000 U/l were observed when Lentinus crinitus grown in agroindustrial byproducts (coffee husks) was used for efficient decolorization of remazol brilliant blue R (Almeida et al., 2018). Moreover, promising growth and production of all three ligninolytic enzymes, namely manganese peroxidase, manganese-independent peroxidase, and laccase, for dye decolorization were also noted with Phanerochaete chrysosporium supplemented with wheat straw as the pre-eminent lignocellulosic substrate (Koyani et al., 2013).
Supplementing the decolorization medium with urea and yeast extract as nitrogen sources supported the decolorization of methylene blue by R. pyridinivorans strain UCC 0003, yielding methylene blue-decolorizing activity of 69 ± 1% and 50 ± 2%, respectively. This finding was consistent with the results of Zhou et al. (2014), who reported enhanced biodegradation of methyl red in the presence of complex organic nitrogen sources such as peptone, yeast extract, and beef extract in comparison to simple organic and inorganic nitrogen sources. Similarly, the use of NH4Cl, (NH4)2SO4, and NH4Cl did not support decolorization of methylene blue by this local isolate, showing less than 25% methylene blue removal after 2 h of incubation period.
Effect of inoculum concentration
There is a direct relationship between the inoculum size and the extent of decolorization of the dye, and therefore, the effect of the inoculum size on dye decolorization is an essential factor that needs to be evaluated. In the present study, the effect of different inoculum concentrations ranging from 4% (v/v) to 20% (v/v) on the rate of methylene blue decolorization by R. pyridinivorans strain UCC 0003 was investigated, and the results are presented in Table 1.
The methylene blue-decolorizing activity increased from 63 ± 0% to 99 ± 1% when the concentration of the inoculum was increased from 4% (v/v) to 16% (v/v) after an incubation period of 90 min. The increase in methylene blue decolorization rate most likely occurred because of the presence of young and active cells in the inoculum (Balraj et al., 2016). Generally, it is expected that as the concentration of the inoculum increases, the rate of methylene blue decolorization should also increase as the growth of R. pyridinivorans strain UCC 0003 becomes faster (Koyani et al., 2013). However, a further increase in the concentration of inoculum from 16% (v/v) to 20% (v/v) did not significantly affect methylene blue removal, resulting in 99 ± 1% decolorization (P > 0.05) as supported by findings reported by Bharti et al. (2019). Consequently, the inoculum concentration of 16% (v/v) was found to be the optimum amount of inoculum for the decolorization of methylene blue.
Effect of the initial concentration of methylene blue
Figure 2A shows the decolorization of methylene blue by R. pyridinivorans strain UCC 0003 at 30°C and pH 6 for 7 h; this experiment was performed to assess the effect of different concentrations of the colorant ranging from 0.1 to 2 g/l on the decolorization performance of the local isolate. A nonsignificant loss of methylene blue was observed in the control group throughout the experimental process, indicating that the reduction of methylene blue concentration in the medium containing active cells of R. pyridinivorans strain UCC 0003 was mainly due to the biodecolorization process. Figure 2A shows that a lower methylene bluedecolorizing activity was observed at a higher concentration of methylene blue than at a lower concentration of the colorant. The methylene blue-decolorizing activity was 56 ± 1% after an incubation period of 7 h at the dye concentration of 2 g/l, while the activity was reduced to 38% at the dye concentration of 1.5 g/l for 7 h. This may be due to the lack of an average attack on each dye molecule by the enzyme that decolorizes methylene blue at high concentrations of dye. This resulted in a decrease in the removal rate of methylene blue. (Zeng et al., 2015). Moreover, the reduced removal of methylene blue in this case may be due to substrate inhibition cause by the blocking of active sites of methylene blue-decolorizing enzymes by the methylene blue substrate (Eslami et al., 2017). In addition, the presence of antibacterial properties along with a high toxicity effect can be observed at high concentrations of methylene blue, and this may have a negative effect on the function of R. pyridinivorans strain UCC 003 as a biocatalyst performing decolorization of the dye (Noraini et al., 2012).
Fig. 2
Decolorization of methylene blue by R. pyridinivorans strain UCC 0003: A) effect of different initial concentrations and B) repeated decolorization of 0.75 g/l methylene blue; star (*) indicates the start of the second cycle; the control was established in the absence of R. pyridinivorans strain UCC 0003; the percentage of methylene blue removal represents mean values of triplicate samples ± standard errors. Incubation was performed at 30°C and pH 6; banana peel was supplemented as the carbon source, and the experiment was performed under static conditions; error bars represent standard error between three determinations
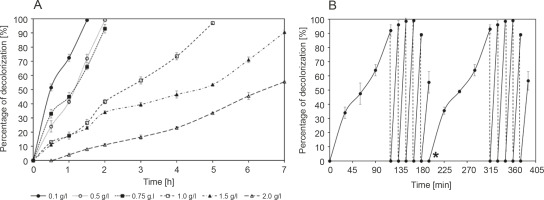
A previous report (Noraini et al., 2012) showed the decolorization of methylene blue at the concentration of only 1 g/l. This study used an isolate detected from a closed drainage system (CDS) located in Prai Industrial Area of Penang, Malaysia. The isolate identified as Sphingomonas paucimobilis could decolorize 85% of 1 g/l methylene blue after an incubation period of 5 days (Noraini et al., 2012). In addition, Zeng et al. (2015) reported a bacterial strain identified as Phanerochaete chrysosporium, which could decolorize 50% of 0.6 g/l after an incubation period of 22 days. Cells of A. faecalis immobilized on biochar as a packing medium exhibited the maximum methylene blue removal efficiency of 35% when subjected to decolorization of 500 ppm of the colorant. This result was achieved due to the presence of the immobilization matrix that provided a protective barrier against the high toxicity of the methylene blue dye and by providing an active target site for bacterial cells to attack the dye molecules (Bharti et al., 2019). Therefore, in the present study, it was interesting to observe that a successful methylene blue-decolorizing activity of 91 ± 2% and 56 ± 1% was achieved when R. pyridinivorans strain UCC 0003 was used as a biocatalyst for the decolorization of 1.5 g/l and 2 g/l methylene blue. To the best of our knowledge, this is the first demonstration indicating the tolerance and decolorization of such a high concentration of dye by a Rhodococcus strain.
A complete removal of methylene blue was observed after 2 h of incubation period when R. pyridinivorans strain UCC 0003 was used as a biocatalyst to decolorize the dye at lower concentrations ranging from 0.1 to 0.75 g/l. However, subsequent addition of the dye to the concentration of 1 g/l led to complete decolorization after 5 h. Collectively, the findings of the present study highlighted the ability of the local isolate to decolorize a wide range of methylene blue concentrations while successfully withstanding the harmful effects of the colorant at higher concentrations, thus making R. pyridinivorans strain UCC 003 a potential biocatalyst candidate for the actual bioremediation of wastewater containing methylene blue.
The capability of R. pyridinivorans strain UCC 0003 to be reused as a biocatalyst for a repeated addition of 0.75 g/l methylene blue dye aliquot under the static condition was also tested in the present study. After the complete removal of the first aliquot of 0.75 g/l methylene blue, which occurred after 2 h of incubation period, another aliquot of 0.75 g/l methylene blue was added to the production medium. The time taken for successful decolorization of methylene blue was reduced by 88% when fresh 0.75 g/l methylene blue was added as the second aliquot, as shown in Figure 2B. An almost complete decolorization of methylene blue was observed upon four subsequent additions of fresh 0.75 g/l methylene blue, which was accomplished within 15 min of incubation period. Compared to the initial inoculation, the removal rate of methylene blue was found to be significantly improved, which indicated that the adaptability of the strain to the colorant had increased and it lasted for four subsequent cycles (Olukanni et al., 2010). However, the fifth addition of fresh 0.75 g/l methylene blue deteriorated the decolorization activity to 56 ± 8%, indicating that the depletion of nutrients may have occurred in the culture medium (Saratale et al., 2009). When the same protocol was duplicated to study the reproducibility of this experimental method, a similar trend was observed; the decolorizing activity was restored to its initial performance when the culture medium was supplemented with a fresh medium after the fifth addition of a fresh aliquot of 0.75 g/l dye. Decolorization of methyl red by Rhodococcus strain UCC 0016 showed a similar trend of the rapid removal of 0.5 g/l of the colorant during the second, third, and fourth additions of fresh aliquots to the culture media under static conditions (Nallapan Maniyam et al., 2020b). The addition of the fifth aliquot of the colorant led to a reduced rate of methyl red decolorization to 57%, and the colorant removal activity was restored during the sixth cycle after the addition of a fresh medium. The percentage of methyl red decolorization was maintained over 90% in the next four cycles, and a similar trend in the experimental results was observed in the present study. Thus, from an economic point of view, R. pyridinivorans strain UCC 0003 could serve as a biological tool for multiple decolorization cycles.
Kinetic study
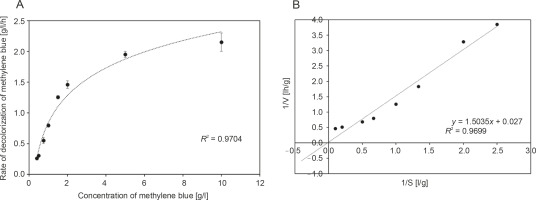
Because the decolorization of a dye is primarily an extracellular enzymatic process, further experiments were performed to establish the kinetics of methylene blue decolorization. The rate of disappearance of methylene blue by R. pyridinivorans strain UCC 0003 at various initial dye concentrations ranging from 0.4 to 10 g/l is shown in Figure 3A. Experimental studies were performed at the optimal pH and temperature under static conditions in order to determine the maximum rate of decolorization of methylene blue and the maximum concentration of methylene blue tolerated by R. pyridinivorans strain UCC 0003. As illustrated in Figure 3A, the increase in the initial dye concentration in the range of 0.4 to 5 g/l yielded an increased rate of decolorization of methylene blue by the Rhodococcus strain. In addition, the catalytic decolorization of methylene blue occurred at a rapid removal rate as indicated by Vmax of 37.04 g/l/h (Handayani et al., 2011). However, a potential occurrence of enzyme-substrate saturation was observed as there was no further increase in the decolorization activity with an apparent Km value of 55.69 g/l for R. pyridinivorans strain UCC 0003, when the concentration of methylene blue was increased to 10 g/l (the inhibition constant Ki can only be calculated if an inhibitor was introduced into the system). Nevertheless, the findings from the kinetic study (Fig. 3A and Fig. 3B) implied that R. pyridinivorans strain UCC 003 could tolerate up to 5 g/l of methylene blue, which is advantageous and promotes the use of this isolate as a biocatalyst in the decolorization of methylene blue on a large scale.
Fig. 3
Kinetics of methylene blue decolorization by R. pyridinivorans strain UCC 0003: A) Michaelis-Menten kinetic model and B) Lineweaver-Burk plot; data represent means of triplicate samples ± standard errors; the rate of disappearance of methylene blue (0.4 to 10 g/l) was measured after a suitable period of incubation in sealed vials at 30°C and pH 6 under static conditions; error bars represent standard error between three determinations
Phytotoxicity study
Methylene blue solution, solution containing methylene blue decolorization byproducts, and distilled water were used to grow the seeds of V. radiata and T. aestivum, and the results are presented in Table 2. After complete decolorization of methylene blue, the decolorized solution was centrifuged at 13 000 × g for 15 min at 4°C. Ethyl acetate was added to the decolorized solution to extract the degradation products by using a rotary evaporator. The mixture was dried and dissolved in distilled water in a sufficient amount to prepare a solution with a final concentration of 0.2 g/l. Comparable findings were reported for seeds grown in solution containing methylene blue decolorization metabolites and seeds grown in distilled water for all the tested parameters, namely germination, plumule length, and radical length (Table 2). This indicates that the decolorized byproducts were nontoxic in nature. Thus, the treated methylene blue solution is nontoxic and could be used for field irrigation application. Treated textile water was used in wheat irrigation and showed better results in terms of yield and nutritional values, as reported previously (Sen et al., 2019). In contrast, the germination of V. radiata seeds in the untreated methylene blue solution was severely inhibited, with a yield of only 48 ± 1%. T. aestivum showed similar results of 50 ± 0% germination. Collectively, the phytotoxicity study revealed that R. pyridinivorans strain UCC 003 was successful as a green biocatalyst to eliminate the toxicity of methylene blue and thus could be capitalized for treating actual wastewaters containing this textile dye.
Table 2
Phytotoxicity studies of methylene blue and its degradation metabolites against Vigna radiata and Triticum aestivum
[i] The experiment was performed for 7 days at room temperature with 0.2 g/l methylene blue and its decolorization metabolites; germination, plumule length, and radicle length are expressed as means of triplicate samples ± standard errors; statistically significant differences (P < 0.05) were observed among the parameters studied between methylene blue solution and its extracted metabolites; water sample was used as a control
Conclusions
Methylene blue is a highly hazardous dye and exhibits recalcitrant properties toward biodegradation; therefore, there is an urgent need to screen for more methylene blue-degrading microorganisms. The genus Rhodococcus, in particular, shows many commercially interesting characteristics such as robustness and diverse metabolic activities, thereby making it an attractive and valuable biological tool in the field of bioremediation and biotechnology. Therefore, the present study used R. pyridinivorans strain UCC 0003 isolated from a sludge sample in Peninsular Malaysia as a biocatalyst to decolorize methylene blue. This isolate exhibited an excellent ability to decolorize methylene blue, with distinct benefits such as the ability to degrade methylene blue over a broad range of temperature and pH values; this aspect will be advantageous for treating wastewater containing the colorant. The complete lack of expensive and harmful chemical substances to aid the decolorization process, and most importantly, the absence of any toxic byproducts of the decolorization process make this approach cost-effective and safe. In addition, the biocatalyst can be repeatedly used, which is beneficial economically. It was interesting to learn that the isolate that was previously not exposed to methylene blue could function as a biological tool to remove the colorant, thereby indicating the pre-eminence of Malaysian biodiversity. The present study also attempted to convert waste to wealth by successfully using agricultural waste to support the growth of R. pyridinivorans strain UCC 0003 during methylene blue decolorization. In future studies, R. pyridinivorans strain UCC 0003 will be immobilized on various support matrices and optimized for use in the bioremediation of actual textile wastewater.







