Introduction
The increase in the human population and the necessity for food production have direct effects on global warming as well as carbon and water footprint. Production and consumption of food result in huge amounts of food waste that needs to be managed. Food waste is primarily generated by agricultural, municipal, wood, food, and other industries, as well as by households (Millati et al., 2019). According to the Food Waste Index Report, households are the primary producers of food waste (61% of total food waste production), followed by food services (26%) and retail (13%) (United Nations Environment Programme, 2021). This scenario presents financial and environmental issues. Increasing interests in environmental protection and circular economy encourage the use of food biodegradable fraction waste as the composting material. In contrast to burying or dumping food waste in landfills of food waste, composting provides an effective food waste management strategy to obtain valuable organic fertilizers. In comparison with artificial fertilizers, compost provides safer nutrients for soil recultivation and is less expensive (Wang et al., 2018).
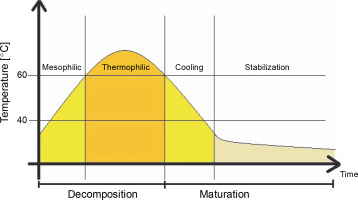
In composting, organic matter is decomposed and stabilized by heterogeneous consortia of microorganisms. During this aerobic process, organic matter is humified into stable organic humate products with the production of heat, water, and carbon dioxide. The process temperature changes depending on the stage, from the mesophilic to thermophilic phase, where the temperature range ranges from below 40°C to 50–60°C or higher (Guo et al., 2012; Awasthi et al., 2017). Temperature changes during the composting process are shown in Figure 1.
Fig. 1
Temperature distribution during the composting process (according to Nikoloudakis et al., 2018)
With changes in temperature, biodiversity and structure of key microbial communities in the composting material change as well (Wei et al., 2018). The primary components of domestic compost are leaves, leftovers of plants, vegetables, coffee grounds, eggshells, and dry grass, which makes compost rich in lignocellulose (Chen et al., 2021). Lignocellulose is complex plant biomass mainly composed of lignin (the outer layer of plants), cellulose, and hemicellulose, which are carbon sources for cellulolytic microorganisms. Cellulose is the primary carbohydrate source in plant cells; however, the presence of lignin, its physical barrier structure, and the crystalline structure of cellulose make the degradation and transformation of the organic fraction difficult (Wu et al., 2017; Lestari et al., 2020).
Enzymatic decomposition of cellulose may be conducted by microbial communities present in the environment, including bacteria, such as Corynebacterium, Aeromonas, Clostridium thermocellum, Cellulomonas spp., and Actinomycetes, and fungi, such as Penicillium sp., Fusarium roseum, Aspergillus niger, and Trichoderma reseei (Gautam et al., 2012; Kogo et al., 2017; Kyaw et al., 2018). Such microbial groups differ from each other in their enzymatic activity, nutritional requirements, and mechanism of growth, i.e., in culture media containing cellulose, where they may show little or no enzymatic activity (Begum and Absar, 2009). The presence and diversity of microbial groups, which are crucial in organic material decomposition, result in better quality of mature compost. Thus, the optimal composition of inoculating microorganisms and the presence of specialized microorganisms may increase the effectiveness of the composting process as they can stabilize the polymeric organic matter and transform it into humic substances (humic acids (HAs), fulvic acids, and humins) (Wu et al., 2017; Zhao et al., 2017).
Therefore, in the present study, autochthonous microorganisms with cellulolytic abilities isolated from compost were used to inoculate composting material. Their effectiveness as a biovaccine and improvement tool in the composting process was evaluated. No simultaneous modifications of the physicochemical parameters of the compost were carried out, i.e., temperature, pH, carbon, and nitrogen concentration. The physicochemical parameters of the inoculated compost were compared with those of the compost not supported by biopreparation. Polymerase chain reaction-denaturing gradient gel electrophoresis (PCR–DGGE), a method that allows monitoring genotype changes (Palaniveloo et al., 2020), was used to determine the genotype structure based on differences in the chosen genome sequences and changes in the community.
Materials and methods
Isolation and identification of cellulolytic bacteria
For the isolation of cellulolytic microorganisms, 1 l of the material was gathered from the one-third depth of the composter with a volume of 600 l. To isolate cellulolytic bacteria, 5 g of the 3-month-old compost was suspended in 45 ml of sterile saline and shaken for 5 min at 120 rpm at room temperature. The samples were left for sedimentation, and then, 1 ml of the resulting supernatant was used to prepare serial dilutions in the range of 1–7. Dilutions of the microbial suspension were prepared in sterile saline and streaked on the medium plates (NaNO3 – 1 g/l, KH2PO4 – 1 g/l, KCl 1 g/l, MgSO4 – 0.5 g/l, yeast extract – 0.5 g/l, glucose – 1 g/l, agar agar – 17 g/l) containing Congo red (1 g/l) and carboxymethyl cellulose (CMC – 5 g/l). To isolate the pure cultures of cellulolytic microorganisms, the plates were first incubated for 24 h at 37°C and then for 6 days at 20°C. After incubation, the plates were covered with 1 M HCl for 15 min, which changed Congo red’s color to blue–violet and inhibited the enzymatic activity of microorganisms (Sazci et al., 1986). Colonies that caused discoloration of the medium were considered having cellulolytic abilities.
The morphology of the isolated cellulolytic bacteria was tested using Gram staining. A smear of bacteria was distributed on a glass slide, air-dried, and fixed in the burner flame. Then, it was stained with Crystal Violet (2 min) and iodine solution (1 min), rinsed with ethanol (30 s), and counterstained with safranin (1 min). The slide was examined under oil immersion at 100 × magnification using a light microscope. To identify strains via 16S rRNA coding gene sequencing, their DNA was isolated using DNA Genomic Mini (A&A Biotechnology, Poland) in accordance with the manufacturer’s instructions. Then, PCR was carried out with the primer pair 27f (5′−AGAGTTTGATCMTGGCTCAG−3′) and 1492 r (5′–TACGGYTACCTTGTTACGACTT–3′) to amplify the 16S rRNA coding gene of the isolate (Wang et al., 2013). Initial DNA denaturation was performed at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 45 s, annealing at 54°C for 45 s, elongation at 72°C for 45 s, and then final elongation at 72°C for 5 min. PCR products were purified from reaction substrates using the Clean-up Kit (A&A Biotechnology, Poland). This purified DNA was sequenced by a chain termination method using a Big Dye Terminator, version 3.1, reagent set (Applied Biosystems Life Technologies, ThermoFisher) and a capillary sequencer 3730 xl DNA Analyzer (Applied Biosystems Life Technologies, ThermoFisher). The obtained sequences were edited and assembled using the Chromas Lite program (Technelysium). They were compared for similar nucleotide sequences using the BLAST (Basic Local Alignment Search Tool) search of the National Center of Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Experiment settings and sampling
The composting process was carried out on the domestic composter in Gliwice, Poland, treated with biopreparation and not subjected to any treatment (control). The material used in composting consisted of garden and household waste (grass, fruits, and vegetables). The mixture was composted in two completely filled rectangular plastic containers (600 l volume, 1.5 m width, 2 m length, 1 m height), inoculated with autochthonous cellulolytic bacteria and control, and stirred once a week while keeping its humidity constant. The experiment was conducted outside when the mean temperature was around 22°C. The temperature of the compost (in the middle and the corners of the container, from where the samples were collected) and ambient temperature were measured once a week (Fig. 2). For inoculation, agar broth slants with isolated cellulolytic bacterial cultures were prepared. After 24 h of incubation at 37°C, the bacteria were added to 500 ml of sterile liquid broth (meat extract 0.4 g/l, peptone 4 g/l, NaCl 3.5 g/l, peptone K 5.4 g/l, yeast extract 1.7 g/l), and the culture was incubated for 48 h at 37°C. The compost in the container subjected to biopreparation was inoculated with three cellulolytic bacteria (Bacillus licheniformis, Bacillus altitudinis, and Lysinibacillus xylanilyticus ) by spreading 500 ml of the bullion liquid medium (BTL, Poland) containing isolated microorganisms (the first day of the experiment) and mixed precisely. The CFU/ml (colonyforming unit) of the biovaccine was not measured since only the response to inoculation was tested in this study, not the content of the added microorganisms.
Fig. 2
Changes in temperature in the middle and the corners of the control composter and the composter inoculated with cellulolytic bacteria during 96 days of the experiment conducted at room temperature
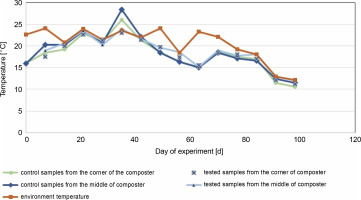
The composting process started in June and lasted for the next 96 days with the addition of unmeasured fresh composting material on the 61st day (in August).
The samples for physicochemical and microbiological tests were collected in sterile Falcon tubes (50 ml) at 2-week intervals, from both containers at two sampling sites: the corners of the control container (CC) and experimental container (EC), and the middle of the CC (CM) and EC (EM) (Fig. 1).
Measurement of physicochemical parameters
The temperature of the composting material and the surrounding environment was measured weekly. Every 2 weeks, the collected samples were analyzed for humidity and the content of organic carbon, total nitrogen, and HAs. The organic carbon content was evaluated using Turin’s method, and the total nitrogen content was determined using the Kjeldahl method (PN-EN 13654-1 : 2002).
Humidity was determined as follows:
where mb is the mass before drying and ma is the mass after drying.
The HA content was evaluated by drying 0.5 ± 0.02 g of the samples at 105°C (72 h) and then adding a mixed equal solution of NaOH (at the initial concentration of 0.1 M) and Na4P2O7 (at the initial concentration of 0.1 M). The compost samples were flooded with the solution in a ratio of 1 : 10 and incubated for 24 h. The samples were incubated for 24 h at room temperature and centrifuged (10 min, 5000 rpm). The supernatants were taken in previously weighed tubes and then acidulated by adding HCl (38%) till their pH reached 1. The samples were centrifuged again (10 min, 4500 rpm), and the supernatants were collected and left to dry at room temperature to rich constant weight. The HA content was calculated as the percentage of the obtained dry precipitate mass in the total mass of a sample:
where mc is the mass of the sample after drying and md is the mass of the sample used for the test.
Culture-based methods
The number of psychrophilic, mesophilic, and sporeforming bacteria was determined on a glucose-enriched solid broth (beef extract 3.0 g/l, peptone 10.0 g/l, glucose 1.0 g/l, sodium chloride 5.0 g/l, agar 15.0 g/l). The number of fungi on Czapek-Dox agar (agar 15.0 g/l, dipotassium hydrogen phosphate 1.0 g/l, iron (II) sulfate heptahydrate 0.01 g/l, magnesium sulfate heptahydrate 0.5 g/l, potassium chloride 0.5 g/l, sodium nitrate 3.0 g/l, and sucrose 30.0 g/l) and the number of Actinomycetes on the medium with nystatin (malt extract 8 g/l, yeast extract 4 g/l, glucose 4 g/l, agar 20 g/l, nystatin 0.05 g/l, and nalidixic acid 0.01 g/l) were also determined. The cultures were incubated for 2 weeks at 20°C for Actinomycetes ; for 24 h at 20 and 37°C for psychrophilic and mesophilic bacteria, respectively; and for 72 h at 20°C for fungi. Spore-forming bacteria were incubated after pasteurization of the diluted compost material at 80°C for 20 min. Then, bacteriological material was spread onto broth plates and incubated for 48 h at 20°C.
PCR–DGGE analysis of the bacterial and fungal communities in containers
DNA from the experimental (three samples collected from the middle and three from the corners of the container) and control (three samples collected from the middle and three from the corners of the container) compost samples was isolated using a previously described mechanical method (Ziembińska-Buczyńska et al., 2015). The filtrate contained DNA and was purified using the Anty-Inhibitor Kit (A&A Biotechnology) to remove HAs and common PCR inhibitors from the samples, in accordance with the manufacturer’s instructions.
To analyze the changes in bacterial and fungal biocenosis during composting, the PCR–DGGE method was used. PCR was carried out to amplify the selected parts of DNA, and the products were subjected to DGGE. PCR amplification targeting the bacterial 16S rRNA coding gene was carried out using the 968F-GC and 1401R primers (Kubartová et al., 2007; Ding et al., 2012). PCR amplification was carried out to detect fungi using the NS1F-GC and FUNG 518R primers (Table 1, Evans and Seviour, 2011; May et al., 2001). PCR was carried out in 30 μl mixture of 1.5 U GoTAQ Polymerase (Promega), 1 × buffer with 2 mM MgCl2, 5 pmol/μl of each primer, and 20 pmol/μl of dNTPs, in a concentration of 0.2 μg/μl. Amplification was carried out in the T-1000 thermocycler (Bio-Rad) according to the parameters described in Table 2. The products obtained were visualized on 1% agarose gel stained with ethidium bromide (0.01 μl/ml, Promega) under UV (ultraviolet) light.
Table 1
Primers used in the study
| Primer* | Sequence (5′ to 3′) | Target | References |
|---|---|---|---|
| 968F-GC | AACGCGAAGAACCTTAC | bacterial 16S rRNA coding gene | Kubartová et al., 2007; Ding et al., 2012 |
| 1401R | CGGTGTGTACAAGACCC | ||
| NS1F-GC | GTAGTCATATGCTTGTCTC | fungal 18S rRNA coding gene | Evans and Seviour, 2011; May et al., 2001 |
| FUNG 518R | ATTACCGCGGCTGCTTG |
Table 2
PCR programs used in the study
The DGGE of PCR products was carried out in a Dcode Universal Mutation Detection System (BioRad) using 8% polyacrylamide gels (37.5 : 1 acrylamide/bisacrylamide, Fluka) with a gradient of 30–60% the denaturant (urea). The gels were run: with bacterial DNA at 40 V for 16 h and fungal DNA at 120 V for 6 h. All gels were run at a constant temperature of 60°C in a 1 × TAE buffer. After electrophoresis, the gels were stained with ethidium bromide (10 mg/μl) for 30 min and destained with MilliQ water for 20 min. The gels were then visualized under UV light and photographed with the Quantity One 1D software (BioRad). This software was also used in the densitometric analysis of gels, which allowed the calculation of the Shannon biodiversity index using the following equation:
where R is the richness of the community and pi is the proportion of individual microorganisms of the i th species in the whole community.
Statistical analysis
The physicochemical and microbial data obtained in this study were expressed as mean values of three technical replicates. Data normality was evaluated using the Shapiro–Wilk test, and Leven’s test was used to identify the equality of the variances. For each parameter, analysis of variance was used to find out whether there was a significant difference between the groups. For normally distributed data, ANOVA was used, and for non-normally distributed data, the Kruskal–Wallis test was used.
Results and discussion
Physical and chemical analysis of composting material
In the control and test samples collected from the middle of the composter, the temperature increased in the first days from about 16°C to a maximum of 28.4°C on the 33rd day for control samples and 23.5°C for the test composter (Fig. 2). The ambient temperature was equal to or higher than the temperature of the samples for almost all testing days, except for the 33rd day for the control composter, when the compost temperature was higher than the ambient temperature (22 ± 2°C for 33 days of the experiment, which decreased below 20°C after that day). The compost temperature was directly related to the ambient temperature, and until the 33rd day, the compost temperature inoculated with microorganisms was lower by 0.5°C than in the control tank. After the 33rd day, the temperature in the control composter was lower by about 1°C. A similar situation was observed in the test and control samples collected from the corners of the composter. The maximum temperature obtained for the control and test samples was about 26°C and about 23°C, respectively. In samples taken from the center and corners of the composter, the temperature of the test samples was equal to or lower than the ambient temperature on all days. These results suggest that inoculation with microorganisms neither decreases nor increases the temperature, which is also confirmed in the literature (Nair and Okamitsu, 2010). However, Rastogi et al. (2019) reported that in composting municipal solid waste with cow dung slurry and additional bacteria, the temperature was higher than in other containers (63°C). For food waste, an extensive increase in the temperature is expected due to the rapid decomposition with the release of high energy as heat (Wang et al., 2018; Awasthi et al., 2018). In the present study, no increased rate of temperature was observed. The inoculation of the composting material with cellulolytic microorganisms was carried out a month after the beginning of the composting process, so the thermophilic phase of composting was not observed in this experiment. Based on the results presented in Table 4, on day 0, after the inoculation of the compost with the biovaccine, the number of psychrophilic bacteria was uncountable, which suggests that a stabilization phase was reached. Thus, probably the thermophilic phase of composting occurred before the inoculation of the composting material with microorganisms. The results of this study are consistent with those of Nair and Okamitsu (2010), who observed no significant temperature differences between control and test samples after inoculating compost material. However, Rastogi et al. (2019) reported that inoculation with microorganisms increases the oxidation reactions by thermophilic microbes, which in turn increases the temperature. It is worth mentioning that the low temperature of the compost might have been due to low oxygen concentrations or reduced concentrations of nutrients (shown by a nonoptimal C : N ratio, which should be between 15 : 1 and 30 : 1; 20–40% (Garg and Tothill, 2009)).
Table 3
Microorganisms identified via Chromas software
| The strain number | Microorganisms from the NCBI database | Percentage of sequence alignment [%] | Accession number | Description |
|---|---|---|---|---|
| 1 | Lysinibacillus xylanilyticus, isolate KWW 56 | 100 | LK391646 1 | gram-positive bacteria isolated from soil and forest humus, capable of xylan hydrolysis (Lee et al., 2010) |
| Lysinibacillus xylanilyticus, isolate KWW 55 | 100 | LK391645 1 | ||
| Lysinibacillus macroides, strain WS26 | 100 | KJ452163 1 | gram-positive bacteria, isolated from cow faeces (Coorevits et al., 2012) | |
| 2 | Bacillus licheniformis strain DGC | 98 | KF840409 1 | gram-positive, mesophilic, soil spore-producing bacteria as well as antibiotics and numerous enzymes, including polysaccharides degrading (Asgher et al., 2020; Jenny Angel et al., 2018) |
| Bacillus licheniformis strain D43 | 98 | KC441778 1 | ||
| Bacillus sp. HL61 1 | 98 | KC705301 1 | gram-positive bacteria isolated from hot springs with high salinity (El-Gayar et al., 2017; Aanniz et al., 2015) | |
| 3 | Bacillus altitudinis strain ML-22 | 100 | KM083101 1 | gram-positive bacteria present in the soil that produce numerous enzymes, including proteases and β-1,3-1,4-glucanase (Tipre et al., 2015) |
| Bacillus altitudinis strain 4BS5 | 100 | KM015453 1 | ||
| Bacillus sp. XQW1 | 100 | KM224522 1 | gram-positive bacteria of the genus Bacillus (Kumar et al., 2020) | |
| 4 | Bacillus licheniformis strain DAS-1 | 99 | KF664027 1 | gram-positive, mesophilic spore-producing bacteria as well as antibiotics and numerous enzymes, mainly proteases; they also secrete enzymes that break down polysaccharides; they are found in soil (Ghani et al., 2013; Wang et al., 2013) |
| Bacillus licheniformis strain wx1 | 99 | KF963618 1 | ||
| Bacillus licheniformis strain RE9 | 97 | JN411571 1 |
Table 4
An abundance of microbial groups during the experiment
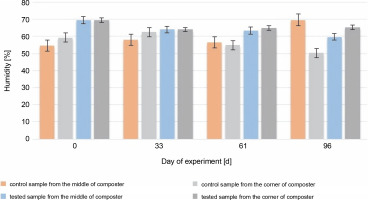
The most accurate humidity for the composted material is between 40 and 60% of the water content. Humidity higher than 60% may cause washing out the nutrients and decrease the volume of air, which will reduce microbial activity (Lestari et al., 2020). In addition, humidity higher than 60% can affect the low C : N ratio (Wang et al., 2018). During most of the experiment, the humidity of the control and test samples (Fig. 3) was optimal (between 50 and 65%).
Fig. 3
Changes in humidity during 96 days of the experiment in the control and test samples collected from the middle and the corners of the composter
Throughout the experiment, changes in the HA content were observed. A higher HA content was measured in the samples collected from the corners of the composter with material inoculated with autochthonous microorganisms (94.2, 20.5, and 8.4% in the compost samples inoculated with microorganisms) than in the control tank (54.4, 5.41, and 9.93%). The samples collected from the middle of the composter inoculated with microorganisms showed a higher concentration of humic substances in the first 61 days (54.3, 15.3, and 8.35%) than the samples collected from the middle of the control composter (37, 9.21, and 62.6%). The HA content of the samples collected from the middle of the control composter was much higher than the rest of the measures (62.6%). In all cases, the HA content was found to be increasing throughout composting, till the 33rd day (from 7.72% in the EM and CM and 49.3% in the EC and CC, to 37% in the CM and 54.4% in the CC and 54.3% in the EM and 94.2% in the EC), which suggests that the maturation phase of the composting has begun 1 month after the inoculation. The increasing HA content confirms that the maturation phase of the compost was achieved. The decrease in humidity measured on the 61st day after inoculation was attributable to the addition of the unmeasured plant biomass and waste to the composting container. New material was added to stage a natural way to use a household composter. This resulted in the mixing of old and new composting material, the start of the new processes of composting, and a decrease in the HA content.
The initial HA content was 7.2% in the middle and 49.3% in the corners of the composter. The organic carbon content was 5.9% in the middle and 8.5% in the corners of the composter. The HA (Fig. 4) and organic carbon (Fig. 5) contents in the compost during the composting process were higher in most of the cases in the experimental composter (HA: 54.3% in the EM, 94.2% in the EC and 15.32% in the EM, 20.52% in the EC; organic carbon: 30.4% in the EM, 30.54% in the EC after 33 and 61 days, respectively) than in the control composter (HA: 37% in the CM, 54.4% in the CC and 9.21% in the CM, 5.41% in the CC; organic carbon: 11.9% in the CM, 25.4% in the CC after 33 and 61 days, respectively) (P > 0.05).
Fig. 4
Changes in the humic acid content during 96 days of the experiment in the control and test samples collected from the middle and the corners of the composter
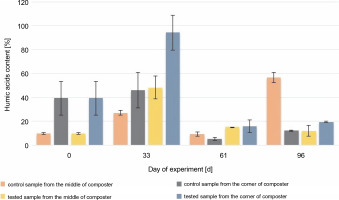
Fig. 5
Changes in the organic carbon content during 96 days of the experiment in the control and experimental samples collected from the middle and the corners of the composter.
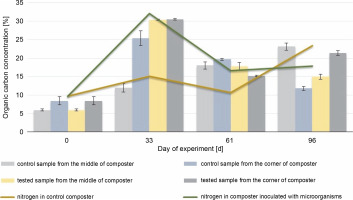
The organic carbon content in the middle and the corners of the composted material after 61 days was higher in the control samples (18% in the CM and 19.7% in the CC) than in the test samples (17.8% in the EM and 15.1% in the EC), which could be due to the presence of the inoculating microorganisms. The highest HA content (94.2%) was observed in the samples collected from the corners of the test composter after the 33rd day, whereas in control samples, the highest HA content (56.5%) was observed after 96 days. HA and organic carbon concentrations influence the maturity of the compost and thus the efficacy of the process. The obtained results suggest that inoculation with microorganisms speeds up the process. The higher the HA content and the lower the organic carbon concentration in the compost, the more mature the compost is (Goyal et al., 2005; Bernal et al., 2009). Based on these results, it can be concluded that the experimental compost reached maturity faster than the control one.
Similar results were reported by Rastogi et al. (2020), who obtained mature compost after 60 days, and Wang et al. (2019) after 35 days of composting. Zhao et al. (2017) observed an increase in the HA content after inoculation with cellulolytic Actinomycetes in comparison with the noninoculated compost. These authors also observed changes in the Actinomycetes community structure. Rastogi et al. (2019), who analyzed the influence of the addition of cellulolytic Bacillus strains, reported a higher degree of humification in composting municipal solid waste supplemented with cow dung slurry or bacteria, in comparison with tanks without any addition. The results obtained in the present study and the literature data indicate that the inoculation of the composting material with microorganisms affects the humification process as a result of the improved degradation of organic substances.
At the beginning of the experiment, the organic carbon and nitrogen content was measured. The initial C : N ratio was 1 : 1.62, and after 33 days, it was 1 : 1.06 in the compost inoculated with microorganisms and 1 : 1.07 in the control tank. Such a low C : N ratio persisted throughout the experiment, and after 96 days of composting, it was 1 : 1.37 in the EM and 1 : 1.01 in the CM. Throughout the composting process, the concentration of nitrogen varied between 10 and 23 mgN/g in control samples and between 10 and 17 mgN/g in experimental samples. However, a comparison of the control and experimental samples showed a higher nitrogen content in the compost inoculated with microorganisms (16.5 mg/l) before adding new composting material (61st day) than in the CC (10.7 mg/l). This was probably attributable to the enhanced decomposition of the plant-based material, which suggests improvements in the composting process. Manu et al. (2017) showed that inoculation with microorganisms improved the degradation of humic and biological substances, i.e., nitrogen, lignin, and polysaccharides, which leads to the humification and production of more stable compounds. On the 61st day, the addition of the new material resulted in a new hard-todecompose material, thus increasing the microbial diversity. After the addition of the new composting material, both organic carbon and nitrogen values were found to be increased (from 12.3% to 13% and 16.5% to 17.8 mg/l in the EC; from 8.9% to 23.1% and 10.7% to 23.4 mg/l in the CC) (P > 0.05). As reported by Rashad et al. (2010), to achieve the best microbial decomposition of organic matter, the C : N ratio should be 32.0 ± 0.92. In the present study, since the beginning of the process, the C : N ratio was 1 : 1.6 (below 1) and was on a similar level throughout the experiment. It may be due to the high concentration of the grass as the composting material, which resulted in a high nitrogen content. A C : N ratio below 1 : 12 is not preferable for the microbial community of the compost (Rashad et al., 2010). Decomposition of the organic matter and mineralization of nitrogen decreased its concentration during the following phases of composting, which shows a decrease in the C : N ratio. In the present study, there was no significant decrease or increase in the C : N ratio. It was also lower than that reported in the majority of the previous studies. These results suggest that under conditions with a lower C : N ratio than reported usually, the composting process may also be effective.
Microbiological analysis
Cellulolytic bacteria isolation and identification
Isolation of cellulolytic bacteria resulted in four bacterial strains that can hydrolyze carboxymethylcellulose. Gram staining showed that all examined strains were gram positive (Fig. 6). The identified strains were able to destain the medium with CMC and Congo red (Fig. 6). Genetic identification was performed using 16S rRNA coding gene sequencing, which showed that three out of four obtained strains belonged to Bacillus sp. (two of them probably to genus B. licheniformis – 98% and 99% match – and one to B. altitudinis – 100% match). The last strain belonged to Lysinibacillus sp. (L. xylanilyticus (100% match)) (Table 3). All identified bacteria are known to be potentially cellulolytic (Awasthi et al., 2015). Mahalik et al. (2018) used the strain of Lysinibacillus sp. in submerged fermentation to produce cellulases. Pan et al. (2012) obtained a stable compost after 75 days of composting in a reactor inoculated with Bacillus subtilis and Pseudomonas sp. and determined that inoculation using a consortium is more effective than using individual isolates. Xu et al. (2019) used B. licheniformis, Aspergillus nidulans, and Aspergillus oryzae in their study. They observed increased efficiency of the composting process after inoculation, which was related to the induced biotransformation of the organic matter by the decomposition of lignocellulose. Moreover, they found that inoculation affects changes in the diversity of microbial community structure. Niu et al. (2022) isolated cellulolytic Bacillus strains from the compost and investigated the influence of inoculation with those strains in short-term composting. After 7 and 14 days, hemicellulose was degraded the fastest in all containers, followed by cellulose and lignin. Moreover, they observed enhanced lignocellulose-related enzyme activity in containers inoculated with Bacillus thermoruber. Li et al. (2020) reported that inoculation with microorganisms (Acinetobacter pittii, B. subtilis subsp. Stercoris, and B. altitudinis) during the cooling stage can increase cellulase activity. Both Li et al. (2020) and Niu et al. (2022) reported a reduction in functional genes related to human diseases and the regulation of microbial community structure.
Abundance and biodiversity of the key functional microbial groups of the composting process and the total bacterial community in the composters
The microbial groups that are abundant in the composting process were mesophilic bacteria, fungi, Actinomycetes, psychrophiles, and spore-forming bacteria. In the samples collected from the compost containers at both the beginning and the end of the experiment, the dominant microorganisms were psychrophilic bacteria (Table 4), which are cold tolerant and can survive in temperatures below 20°C. Their presence was also confirmed during the initiation and maturation phases of composting in a previous study (Wang et al., 2014). After inoculation, the number of the mesophilic bacteria decreased in the CM from 250.8 to 7.335 mln CFU/g and increased in the samples collected from the corners of the CC from 189.6 (CC) mln CFU/g up to an uncountable number. The number of mesophiles in the EC decreased to 3.690 mln CFU/g in the EM and 1.371 mln CFU/g in the EC. After the 33rd day, the number of mesophiles was on a similar level in the container inoculated with the biovaccine (EC and EM) until the end of the experiment, which increased only in the samples of the compost inoculated with microorganisms collected in the middle of the container (to 253.5 mln CFU/g). The number of spore-forming bacteria increased during the composting process from 0.3 mln CFU/g in the EC and EM to 2.7 in the EC and 3.3 mln CFU/g in the EM in August (day 61). At the end of September (the 96th day of the experiment), the number of spore-forming bacteria was 17.7 mln CFU/g in the middle of the compost inoculated with bacteria and 83.7 mln CFU/g in the corners of the inoculated compost. These results show that either ambient or compost temperature does not affect the number of spore-forming bacteria. The increasing number of sporeforming bacteria was probably attributable to the depletion of substrates, which forced some bacteria to produce endospores (Gray et al., 2019).
The number of Actinomycetes steadily increased from 0.6 mln CFU/g in the CM and 0.3 mln CFU/g in the CC. The increase was higher in the corners (843 mln CFU/g in the control and 648 mln CFU/g in the inoculated compost) of the composting container than in the middle (570 mln CFU/g in the control and 232.5 mln CFU/g in the inoculated compost). Actinomycetes can decompose complex compounds such as lignocellulose, so their increased presence confirms the depletion of simple organic compounds and the progressive decomposition of compounds such as cellulose (Wei et al., 2019). Their presence and an increase in their number also underline the maturation process and finalization of the composting process. Furthermore, Abdulla and El-Shatoury (2007) reported that Actinobacteria can decompose lignocellulose, and Zhao et al. (2016) confirmed the increase in cellulose degradation in composting chicken manure, cow dung, pig manure, kitchen waste, and municipal solid waste.
At the beginning of the experiment, the number of fungi was 244.8 mln CFU/g in the EM and 43.2 mln CFU/g in the EC. On the 33rd day, the number of fungi increased in all samples. In the control samples, the number of fungi was 3.763 (CM) and 2.039 mln CFU/g (CC). In the samples inoculated with bacteria, the number of fungi was 6.446 mln CFU/g (EM) and 1.906 mln CFU/g (EC). After the 33rd day, the number of fungi was rapidly increasing and was found to be 173.9 mln CFU/g (CM), 112.9 mln CFU/g (CM), 33.2 mln CFU/g (EM), and 109.2 mln CFU/g. On the 61st day, the number of fungi decreased to 37 mln CFU/g in the control samples and 64 and 24 mln CFU/g in the compost inoculated with microorganisms in the middle and the corners. This decrease in the number of fungi suggests that the compost is matured; thus, after the 33rd day, the maturation phase started (Ma et al., 2019). This is also confirmed by the results of the HA and organic carbon content analysis. Psychrophilic bacteria were the dominant group of microorganisms at the beginning and the end of the composting process, whereas mesophilic bacteria and fungi were less abundant. On the 61st day, the abundance of psychrophilic bacteria decreased in all containers by about 1000 mln CFU/g in comparison with the 33rd day. On the 61st day, the ambient temperature was higher than in containers. The number of psychrophilic bacteria in the CC increased on the 96th day again to an uncountable number in the CM and EM and to 219.6 mln CFU/g (EC) and 16.2 mln CFU/g (EM). The number of mesophilic bacteria was higher in the corners (uncountable on the 33rd day, 1.038.6 mln CFU/g on the 61st day, 1.200 mln CFU/g) and the middle (7.335 mln CFU/g on the 33rd day, 405 mln CFU/g on the 61st day, and 435 mln CFU/g) of the CC than in the container subjected to biopreparation (EC: 1.371 mln CFU/g on the 33rd day, 613 mln CFU/g on the 61st day, 1.290 mln CFU/g on the 96th day; EM: 3.690 mln CFU/g on the 33rd day, 253.5 mln CFU/g on the 61st day, and 153 mln CFU/g on the 96th day). After 61 days, a higher number of mesophilic bacteria were present in the corners of containers than in the middle. A higher number of spore-forming bacteria (above 60 mln CFU/g), Actinomycetes (150.8 mln CFU/g in the middle and 9.5 mln CFU/ml in the corners), and fungi (6.446 mln CFU/g in the middle and 1.906 mln CFU/g in the corners) were observed in the experimental sample after inoculation, but in the rest of the experiment, the number of Actinomycetes was higher in the control samples (about two times higher than in the compost inoculated with bacteria). The number of Actinomycetes was found to be increasing during the test for 61 days, and the highest number was observed on the 61st day in the EC (900 mln CFU/g). Actinomycetes can biodegrade difficult-to-decompose substances such as cellulose. Their increasing number suggests the depletion of easy-to-biodegrade organic substances and the decomposition of multimolecular compounds. On the 33rd and 96th days, spore-forming bacteria were the most abundant (above 60 mln CFU/g in the experimental samples, 44.7 mln CFU/g in the CM and 90 mln CFU/g in the CC, and CM: 570, CC: 843, EM: 232.5 and EC: 648 mln CFU/g). On the day of inoculation, the abundance of all microorganisms decreased and then increased in the following days. The number of spore-forming microorganisms was found to be increased on the 33rd day due to changed conditions. When the abundance of all microorganisms was decreasing, the number of spore-forming bacteria was increasing. Changes in the number of microorganisms occurred faster in the experimental samples than in the control samples. This indicates that inoculation speeds up the composting process, which was further confirmed by the HA and organic matter contents. Similarly, Rastogi et al. (2019) reported that in the container inoculated with four Bacillus strains, the compost matured earlier than in control and other test tanks; moreover, the inoculated compost showed better stability.
PCR-DGGE analysis
PCR amplification and separation using DGGE showed changes in the bacterial community structure in different composting areas. Based on the obtained PCR-DGGE fingerprints, the Shannon biodiversity index (HN) was calculated (Fig. 7 and Fig. 8). The values of the Shannon biodiversity index below 1 suggest low species diversity. No statistically significant difference in the number of microorganisms was observed between test and control samples on the 0th and 33rd days. On the 61st day, a decrease in abundance in the samples collected from the middle of the composter was noticed, which was attributable to the new substrate addition and the adaptation of microorganisms to new conditions. This may be related directly to inoculation and extensive growth of specific autochthonous bacteria.
Fig 7
Shannon Biodiversity Index calculated for bacteria biocenosis in samples collected from the middle and the corner of composters
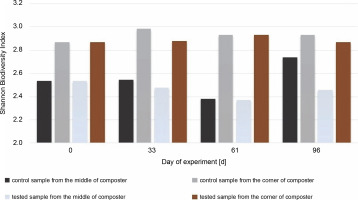
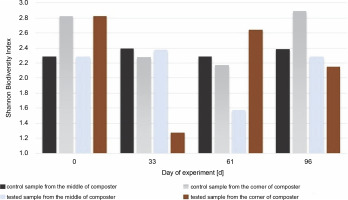
Fig 8
Shannon Biodiversity Index calculated for fungal biocenosis in samples gained from the middle and the corner of composters
On the 96th day, due to the addition of new substrates on the 61st day, the Shannon biodiversity index of microorganisms in the CM increased (from 2.38 to 2.74). Each phase of composting was characterized by specific bacterial composition. Pseudomonas, Bacillus, Cellulomonas, Clostridium, Flavobacterium, etc., were common in the mesophilic phase of composting, whereas Streptomyces and Actinobacteria were dominant in the subsequent phases (Sánchez et al., 2017). It is worth mentioning that many Actinobacteria species are commonly found on plants and straws (Lewin et al., 2016). Therefore, substrate addition may increase the biodiversity of the investigated microorganisms. New microbial groups can be implemented together with the substrate, and the number of free ecological niches for bacteria due to different types of substrate increased.
In the samples collected from the middle of the container inoculated with bacteria, the Shannon diversity index increased to a level close to the one measured on the 33rd day (to H′ = 2.46 from 2.37). This may suggest that bacteria used for inoculation are efficient in producing enzymes and digesting the substrate. There were not many substrates left for the other microorganisms present, and the number of free niches to occupy was low. The number of bacteria in the corners of the inoculated compost did not change much during the test, even after inoculation, and was H′ = 2.86 on the first day and H′ = 2.93 on the 61st day.
The highest value for the Shannon diversity index was observed in the control sample collected from the corners on the 0th and 96th days, H′ = 2.82 and H’= 2.89, respectively. This indicates that the microorganisms in the corners had access to a more diverse range of substrates than those in the middle of the container. The diversity in the corners of the experimental composter decreased on the 33rd day from H′ = 2.82 to 1.28, probably due to the lack of degradable substances in the composter. Furthermore, the fungal biodiversity increased on the 61st day as a possible response to the addition of the new composting material (from H′ = 1.3 to H′ = 2.64). In the experimental samples collected from the middle of the composter on the 61st day, the Shannon biodiversity index was lower (H′ = 1.6) than on other days (around H′ = 2.3). Fungi produce enzymes more effectively than bacteria, and their presence in the composting material improves the degradation of multimolecular compounds. The material used for the composting process was composed of food leftovers and plants; thus, fungi with cellulolytic potential might be present in the waste. Although the presence of microorganisms is dependent on the available substrates, the content of the compost has a significant effect on the fungal community (Xu et al., 2019). With the depletion of the substrates, the fungi abundance decreased, and the addition of new material increased biodiversity. Comparing the Shannon biodiversity index of bacteria and fungi, the highest increases in biodiversity were observed for the fungal community. However, fungi seem to be less fragile or easily adaptable to any changes, which may be attributable to the increase in the Shannon biodiversity index of fungi. These diversity changes may be observed in the samples inoculated with microorganisms in the corners of the composter (33rd day: H′ = 2.9 bacteria index, H′ = 1.3 fungi index; 61st day: H′ = 2.4 bacteria index, H′ = 2.65 fungi index). Importantly, no decrease in the biodiversity of bacteria was observed as a response to the increased number of different fungi. This suggests that those two groups did not affect each other. Xu et al. (2019) reported different results, who observed significant changes in the diversity of microorganisms after inoculation.
Conclusions
Composting is a complex process involving many microorganisms. It is a time-consuming process; thus, methods for its improvement and acceleration are important. The addition of microorganisms is an eco-friendly way to achieve a faster and better decomposition of organic substances to obtain stabilized compost. The findings of the present study showed that inoculation of the compost with four strains of Bacillus can improve the composting process but affect the microbial community already present in the composting material by decreasing the biodiversity, without harmful impact on the process. Further research is needed to investigate the effectiveness of the biovaccine used on different composting materials and determine its exact amount (CFU/g) needed for inoculation.