Introduction
Microorganisms serve as natural resources of bioactive molecules. Among various bioactive entities, microbial pigments have recently gained wide research interest because of their natural origin and safe application. Melanins are one such group of pigments with complex structures and physicochemical properties (Banerjee et al., 2014). Most of the living organisms on the Earth produce melanin to protect themselves from detrimental effects of solar radiation (D’Ischia M et al., 2015). Although many microorganisms can produce melanin, their scaling up and extraction process require further optimization as melanin is produced inside melanosomes (Toledo et al., 2017). Some microorganisms such as Streptomyces, Nocardiopsis, Pseudomonas, Aspergillus, Auricularia, and Armillaria have attracted the attention of scientists because of their ability to produce extracellular melanin which can be easily extracted and purified (Kiran et al., 2014; Raman et al., 2015; Sun et al., 2016; El Naggar and El-Ewasy, 2017; Kiran et al., 2017; Ribera et al., 2019). Jalmi et al. (2012) reported maximum melanin production (6.6 g/l) by the fungus Gliocephalotrichum simplex after 6 days of incubation. On the other hand, Streptomyces kathirae SC-1 produced 13.7 g/l melanin after 5 days of incubation (Guo et al., 2014). Most of these microorganisms depend on the exogenous supply of precursors such as mono-/diphenols for extracellular melanin synthesis (Tran-Lyet al., 2020). Melanin has gained importance because of its potential application in antioxidant and sunscreen products, organic semiconductor devices, biomedical devices, metal chelators, and dyeing agents (Ali Amal et al., 2011; Battistella et al., 2020; Tran-Ly et al., 2020). A study revealed that the thermal stability of the synthetic polymer polymethyl methacrylate was enhanced when eumelanin was added to the mixture. It was shown that melanin remarkably increased the decomposition temperature of the polymer by altering its radical initiated chain scission behavior (Shanmuganathan et al., 2011). A recent report showed the application of melanin as a colorant for soft contact lenses (Ahn et al., 2019). Another study described the use of melanin as a contrast agent in imaging application (Williams, 1994). Melanin extracted from Streptomyces sp. and Pseudomonas balearica shows significant antimicrobial activity against pathogenic bacteria such as Escherichia coli, Staphylococcus aureus, Lactobacillus vulgaris, Vibrio cholera, Proteus miralis, Salmonella typhae, Salmonella paratyphae, and Klebsiella oxytoca (Vasanthabharathi et al., 2011; Zerrad et al., 2014). Because of its unique properties such as heterogeneity, hydrophobicity, and negative charge, the structural analysis of natural melanin is challenging (Pralea et al., 2019).
Many microorganisms can synthesize melanin through multiple pathways. For instance, microorganisms such as Streptomyces glaucescens, Neurospora crassa, and Marinomonas mediterranea can produce melanin by using a pathway similar to that of mammals, which involves the enzyme tyrosinase (Serrano et al., 2004; Kamal et al., 2014; Toledo et al., 2017), while other microorganisms such as Aspergillus fumigatus, Talaromyces marneffei, and Sporothrix schenckii use the DHN pathway/malonyl coenzyme for melanin synthesis (Langfelder et al., 1998; Romero-Martinez et al., 2000; Woo et al., 2010; Cordero and Casadevall, 2017). Certain microorganisms such as Cryptococcus neoformans and Bacillus weihenstephanensis follow the laccase enzyme pathway, while bacteria such as Vibrio cholerae, Hyphomonas sp., and Shewanella colwelliana use the homogentisic acid pathway for melanin synthesis (Kotob et al., 1995; Eisenman et al., 2007; Wang et al., 2011; Drewnowska et al., 2015). Melanins produced through the DHN pathway are localized intracellularly; consequently, they are difficult to purify and hence not desirable for large-scale production (Toledo et al., 2017). Therefore, microorganisms that produce extracellular melanin within a short incubation period are of industrial interest and are relatively economical. An analysis of the melanogenic genes of different taxa would provide better understanding on the importance of their evolutionary aspects (Esposito et al., 2012). In view of this, the present study aimed to screen melanin-producing potential organisms isolated from harsh habitats such as lime quarries in Karnataka, India. The screening process was followed by characterization of melanin and assessment of its antibacterial property. In addition, an attempt was made for comparative analysis of tyrosinase gene, protein, and common central domain (CCD) sequences of selected organisms.
Materials and methods
Screening of soil samples for melanin-producing microorganisms
Eleven soil samples collected from lime quarries in Karnataka, India, were pretreated and initially isolated on starch casein agar medium. The isolates for extracellular production of tyrosinase and melanin were screened by the streak plate culture method on a tyrosine gelatin beef extract (TGB) agar medium containing tyrosine 0.5%, gelatin 0.5%, beef extract 0.3%, agar 2%, and distilled water with pH 8.0. The inoculated plates were incubated at 40°C for 5 days. On the fifth day of incubation, the plates were visually assessed for zones of catalysis, and different intensities of brown color were recorded as positive for the synthesis of extracellular tyrosinase and melanin by the test isolates (Shivaveerakumar et al., 2013).
Identification of the potential isolate
The potential isolate for melanin production was identified preliminarily by Gram staining (Bartholomew and Mittwer, 1952) and motility test by the hanging drop method on a cavity slide. Molecular characterization of the isolate was performed by 16S rRNA gene sequencing using the primers 27F (5′–AGAGTTTGATCMTGGCT CAG–3′) and 1525R (5′–AAGGAGGTGATCCAGCC– 3′) (Thorat and Dastager, 2018). The ability of the bacterial isolate to utilize different simple carbohydrates (0.3% w/v) such as glucose, fructose, sucrose, maltose, raffinose, and xylose as the sole source of carbon was assessed in M9 minimal medium at 37°C for 24 h (Schleheck et al., 2009). The growth of bacteria was determined by measuring the optical density (OD) at 600 nm.
Optimization of physicochemical variables on melanin production
The influence of various factors on melanin production by a potential isolate was studied in L-tyrosine broth incubated at 40°C for 24 h with agitation speed of 120 rpm (Madhusudhan et al., 2014). The effects of pH (6.0–10.0), temperature (30–50°C), and L-tyrosine concentration (0–2 g/l) were studied. The effects of different nitrogen sources (0.5% each of beef extract, peptone, gelatin, yeast extract, and casein), metal salts (0.01% each of CuCl2, CoCl2, CuSO4, MnCl2, and MgCl2), and salinity (0–2% NaCl) on the production of melanin by the potential isolate were also assessed. All the experiments were performed in triplicate, and the results are presented as mean ± standard deviation.
Production and purification of melanin
Submerged production of extracellular melanin by the potential isolate was accomplished in an optimized broth with agitation speed of 120 rpm for 24 h. The production broth was centrifuged after 24 h of incubation to separate the cell mass from other debris. The supernatant obtained was acidified with 1N HCl till it reached pH 2.0 and was allowed to stand for 96 h for melanin aggregation. The aggregate formed was separated and purified by centrifugation at 10 000 rpm for 20 min, followed by washing with ether, ethanol, acetone, and deionized water to remove impurities (Fava et al., 1993). The purified melanin was air-dried and stored at 4°C for further use.
Characterization of melanin
Purified melanin was assessed for its solubility profile in different solvents such as water (pH 7.0), 1N HCl, alkaline solutions (1N NaOH and 1N NH4OH) and organic solvents such as dimethyl sulfoxide (DMSO), dimethyl formamide (DMF), methanol, ethanol, n-butanol, ethyl acetate, acetone, n-hexane, and diethyl ether. Other physical tests such as precipitation of melanin with 1% FeCl3 and bleaching with alkaline H2O2 (10%) were performed as reported previously (Gibson and George, 1998). Melanin was characterized by spectroscopy and chromatographic methods. In brief, melanin was dissolved in 0.1N NaOH and scanned in the range of 200–800 nm UV-Vis spectroscopy. Thin layer chromatography (TLC) was performed by using ethyl acetate and n-hexane (7 : 3) along with 0.1% formic acid as the mobile phase. Highperformance liquid chromatography (HPLC) was performed by dissolving melanin (10 μg) in 10 μl of diluted KOH (0.01N) solution, and the volume was made up to 1 ml with methanol and filtered through 0.2 μm Millex membrane (MF-MilliporeTM). The mobile phase comprised 0.1% formic acid and methanol in the ratio of 85 : 15, and the detection was performed with a C18 KinetexTM 5 μm, 260 × 4.6 mm column at the flow rate of 1 ml/min at 222 nm in the diode array detector (Singh et al., 2018).
Antibacterial activity of melanin
The agar well diffusion assay was performed to assess the antibacterial activity of the melanin extracted from the bacterial isolate against the test pathogenic bacterial species E. coli, P. aeruginosa, Streptococcus entericus, and B. subtilis (Arun et al., 2015). The culture medium of each test isolate incubated for 18 h in nutrient broth was swabbed separately on the surface of sterile nutrient agar plates. Eight wells of 10 mm diameter were punched on each plate and loaded with 100 μl of different concentrations of melanin (300, 250, 200, 150, 100, and 50 μg/well and labeled as A–F, respectively), streptomycin (1 mg/ml), and DMSO. All the plates were incubated at 35°C for 24 h. Streptomycin was used as a positive control, while the solvent DMSO was added as a negative control. The diameter of the zones of inhibition were measured to assess the antibacterial activity.
Comparative analysis of tyrosinase gene and protein sequences from selected organisms
The sequences of the tyrosinase gene from selected organisms were compared and analyzed to find the closest match with the human tyrosinase gene. A total of 72 tyrosinase genes and their reference protein sequences from prokaryotic and eukaryotic organisms (Table 1) were retrieved from the National Center for Biotechnology Information (NCBI) in FASTA format. The reference protein sequences were analyzed to find conserved motifs by using the KEGG’s Motif Search web tool allied with Pfam database. The selected gene sequences, their respective proteins, and conserved motif sequences were separately subjected to multiple alignment using Clustal Omega 1.2.2 in Geneious software 10.2. After the multiple alignment, a distance matrix of sequences and phylogenetic tree were constructed to comprehend the evolutionary relationship between the tyrosinase sequences of selected organisms and Homo sapiens (NM_000372.5) (Esposito et al., 2012). H. sapiens was selected as the out group while constructing the phylogenetic tree.
Table 1
List of selected organisms for the retrieval of tyrosinase gene and protein sequences
Results
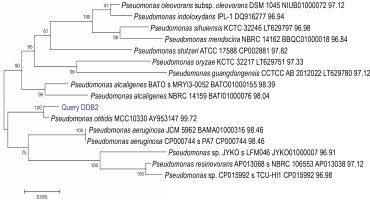
A total of 58 isolates were obtained from lime quarry soil samples collected from Karnataka, India. All the 58 isolates were screened for melanin production, and only six isolates produced dark brown pigment on the TGB agar medium. Among the six positive isolates, the bacterial isolate named DDB2 (D: Name of Research Guide, Prof. Dayanand; D: Name of the Research Student, Deepthi; B: Type of isolate, bacteria; 2: number of the isolate) showed a large zone of pigmentation (38 ± 0.5 mm) after 24 h as compared to the other five isolates (supplementary Table 1), and hence, it was selected for further studies. The isolate DDB2 was gram-negative, motile, rod-shaped bacteria that produced melanin under alkaline conditions. The potential isolate DDB2 was identified by 16S rRNA gene sequencing, and the phylogenetic tree (Fig. 1) was constructed using the neighborjoining method that showed a homology of 99% with Pseudomonas otitidis. The GenBank accession ID of P. otitidis DDB2 is MG585039. Figure 2 shows the extracellular dark brown pigment production by the isolate DDB2 on TGB agar. Carbohydrate utilization test showed that only glucose was utilized by the isolate DDB2, while other carbohydrate sources were not metabolized and no turbidity was observed (supplementary Table 2). The growth of DDB2 on M9 medium with glucose was compared with its growth in nutrient broth; the OD values of 0.24 ± 0.02 and 0.64 ± 0.02 were achieved in the former and latter media, indicating that the isolate poorly fermented sugars but actively utilized complex ingredients of nutrient broth.
Optimization of melanin production
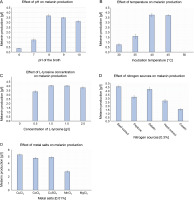
The influence of physicochemical parameters such as pH, temperature, L-tyrosine concentration, metal salts, and salinity on melanin production was assessed. The amount of melanin produced increased with the increase in pH from 6.0 to 8.0, but gradually decreased from pH 9.0 to10.0 (Fig. 3A). Melanin production exponentially increased from 30 to 40°C, and a further increase in temperature to 45°C slightly increased the production of melanin by 0.13 g/l (Fig. 3B). However, the growth of P. otitidis DDB2 was completely inhibited at 50°C. The use of the precursor L-tyrosine resulted in the maximum production of melanin (4.07 ± 0.07 g/l) at the concentration of 1.5 g/l (Fig. 3C). However, the absence of L-tyrosine resulted in no melanin formation (0.0 g/l), and hence, this precursor was found to be mandatory for the synthesis of melanin by P.otitidis DDB2. Among the different nitrogen sources tested, 0.3% of beef extract or gelatin was found to exert a similar positive effect on the amount of melanin production (4.6 ± 0.10 and 4.51 ± 0.10 g/l, respectively). Therefore, either beef extract or gelatin at the concentration of 0.3% was included in the final medium for melanin production. Casein was found to produce the least amount of melanin (1.56 ± 0.10 g/l) than the other nitrogen sources tested (Fig. 3D). The results also showed that 0.01% CuCl2 significantly affects melanin production by P. otitidis DDB2 (Fig. 3E). The metal salts CoCl2 and CuSO4 showed moderate increment in melanin production, whereas MnCl2 lowered melanin production by 2.54 ± 0.12 g/l. Complete inhibition of melanin synthesis was observed with MgCl2. The salinity range of 0–2% neither enhanced nor reduced melanin production. Because the presence of NaCl did not greatly influence the amount of melanin production, it was excluded from the final optimized medium. The optimized conditions for the maximum production of melanin were found to be pH 8.0, incubation temperature 45°C, L-tyrosine (1.5 g/l), beef extract (0.3%), and CuCl2 (0.01%). Melanin production by P. otitidis DDB2 increased from 3.81 ± 0.1 g/l to 5.3 ± 0.1 g/l under the optimized conditions.
Fig. 3
Optimization of process variables for melanin production by P. otitidis DDB2. A) effect of pH on melanin production; B) effect of temperature on melanin production; C) effect of L-tyrosine concentration on melanin production; D) effect of nitrogen sources on melanin production; E) effect of metal salts on melanin production
Characterization of melanin
Purified melanin was soluble in alkaline solutions and in solvents such as DMSO and DMF, but it was insoluble in all the other solvents tested (Table 2). Melanin was precipitated by FeCl3 and turned colorless after reacting with H2O2. The requirement of L-tyrosine as the precursor, the dark brown color, and the solubility profile of the compound revealed that the pigment extracted from P. otitidis DDB2 belonged to the class eumelanin/DOPA melanin.
Table 2
Solubility of melanin in different solvents tested
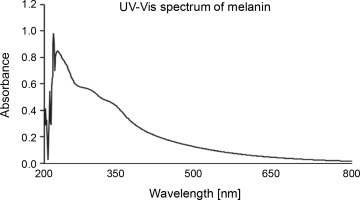
The UV-Vis absorption spectrum of melanin showed the maximum absorbance between the wavelength range of 215–350 nm (Fig. 4). The chromatogram developed on the TLC plate showed a broad spot with the retention factor (Rf) of 0.48. HPLC with 0.1% formic acid and methanol (in the ratio of 85 : 15 ratio) as the mobile phase showed three major peaks with retention time of 1.620, 2.273, and 2.693 min corresponding to 10.01%, 63.20%, and 17.42% of melanin constituents, respectively (supplementary Fig. 1).
Antibacterial activity of melanin
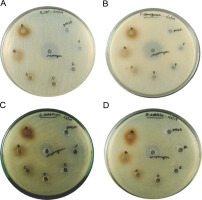
Purified melanin was assessed for its antibacterial activity (Fig. 5). A maximum diameter of 13 ± 2 mm for the zone of inhibition was observed for 300 μg of melanin against S. entericus, whereas the diameter of 12 ± 2 mm was observed for the zones of inhibition with 300 μg of melanin against E. coli, P. aeruginosa, and B. subtilis (supplementary Table 3). The zones of inhibition were 12 ± 1 mm in diameter for 250 μg of melanin against E. coli, P. aeruginosa, and S. entericus. For all the other tested concentrations of melanin, the diameter of zones of inhibition was 11 ± 1 mm against the tested pathogenic bacteria. The zones of inhibition for the positive control streptomycin were 22 ± 1, 24 ± 2, 19 ± 2, and 18 ± 1 mm against E. coli, P. aeruginosa, S. entericus, and B. subtilis, respectively, whereas the negative control DMSO did not show any inhibition zone.
Comparative analysis of tyrosinase gene sequences
Tyrosinase gene sequences of selected organisms retrieved from NCBI were aligned, and a phylogenetic relationship was established to determine the relatedness of the tyrosinase gene of H. sapiens with that of other organisms such as Pseudomonas sp., Streptomyces sp., several fungal species, S. officinalis, Vibrio sp., and tyrosinase-related protein 1 (TYRP1) of H. sapiens. The resultant phylogenetic tree (Fig. 6) showed that the difference in the tyrosinase gene sequences increased in clockwise direction with respect to H. sapiens. The distance matrix was generated by Geneious software for the tyrosinase gene, protein, and CCD sequences, which showed the distances between the sequences of organisms based on percentage identity (%) of the nitrogen bases or amino acid residues (supplementary Table 4).
Comparative analysis of tyrosinase gene sequences of selected organisms with the tyrosinase gene sequence of H. sapiens (NM_000372.5)
Among all the selected organisms, the TYRP1 gene sequence of H. sapiens was found to be more related to the tyrosinase gene sequence of selected H. sapiens with 34.7% identical bases, whereas S. officinalis showed 21.9% and V. nigripulchritudo showed 14.6% identity. Among the selected fungi, T. marneffei ATCC 18224 showed the highest similarity (20.5%) and G. morbida showed the lowest similarity (1.8%) with H. sapiens. Among the selected species of Streptomyces, S. subrutilus and S. fungicidicus showed the maximum (12.4%) and the least (0.6%) percentage of identity, respectively. On the other hand, P. proteolytica and P. frederiksbergensis showed the highest (14.6%) and the least (6%) similarity with the tyrosinase gene sequence of H. sapiens, respectively.
Comparative analysis of tyrosinase protein sequences of selected organisms with the tyrosinase protein sequence of H. sapiens (NP_000363.1)
The TYRP1 sequence of H. sapiens showed 39.3% identity, while those of S. officinalis and V. nigripulchritudo showed 12.3% and 11.9% similarity, respectively, with the tyrosinase protein sequence of H. sapiens. A. niger showed the highest (12.4%) and G. morbida showed the lowest (7.9%) identical amino acid residues among the selected fungi. S. azureus showed the maximum (22.9%) and S. fungicidicus showed the least (3%) percentage of identity, whereas among the selected species of Pseudomonas, P. fluorescens exhibited the highest (20.8%) and P. gingeri showed the least (10.5%) percentage of identity with the tyrosinase protein sequence of H. sapiens.
Comparative analysis of CCD of tyrosinase protein sequences
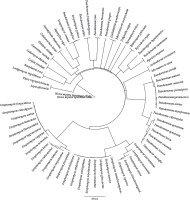
The CCD of the tyrosinase protein sequences of most selected organisms showed 100% identity with the CCD of H. sapiens (supplementary Table 3), and all these organisms exhibited common conserved residues at the positions 164–220 (PDQFRQNIPVLKNEVDRYLTNPSF VRFQADLDRGAHGAIHVFVGGQNSSSPLPANS). The CCD of P. putida, P. chlororaphis, and P. gingeri showed 56.35, 61.4, and 74.2% identity, respectively, with that of H. sapiens, respectively. However, certain species of Streptomyces such as S. zinciresistens, S. pluripotens, S. olivochromogenes, S. virginae, S. canus, S. avermitilis, S. cyaneus, S. violaceoruber, S. katrae, S. fodineus, S. fungicidicus, and S. wuyuanensis and the fungal species G. morbida exhibited no identity with the CCD of human tyrosinase protein sequence. Although these species did not harbor a CCD similar to that of H. sapiens, they were found to possess a conserved amino acid sequence (from position 31 to 73, AAMYEISELAVGDK RGYSALARAHGYDQDLCHDLDWAFLTWHR) that is commonly observed in other microorganisms selected in the present study. The multiple alignment of CCDs of the selected organisms is shown in Figure 7.
Discussion
From preliminary screening, a single isolate named DDB2 showing the largest zone of extracellular melanin production was selected as the potential isolate and used for further studies. The other five isolates that produced maximum zones of pigmentation after 72 h of incubation were excluded from the study because the time of pigment production was too long and the diameter of the zone of pigmentation was smaller than that for the isolate DDB2. The isolate P. otitidis DDB2 could not utilize the carbohydrates provided, except glucose. However, it was observed that DDB2 poorly utilized glucose, but exhibited a rich growth on nutrient agar medium, which indicated the organism’s preference for complex nutrients. The genus Pseudomonas prefers organic acids or amino acids over sugars as their carbon source (Daniels et al., 2010). P. otitidis DDB2 produced the maximum amount of melanin at an alkaline pH of 8.0 (3.81 ± 0.1 g/l) and a high temperature of 45°C (3.85 ± 0.1 g/l). A similar requirement of alkaline pH (9.0) and temperature 45°C was observed for melanin production by Streptomyces sp. isolated from limestone quarries in Deccan traps (Quadri and Agsar, 2012). Previous studies have shown that microorganisms produce melanin under certain stress conditions such as poor nutrient supply, high temperature, or high osmotic pressure (Fogarty and Tobin, 1996; Tran-Ly et al., 2020). This may be the reason for the requirement of high temperature (40 ± 5°C) and high pH ≥ 8.0 for melanin production by the isolated Pseudomonas sp. in this study. The isolate P. otitidis DDB2 showed an obligatory requirement of the precursor L-tyrosine for melanin production. Previous reports show that Pseudomonas stutzeri HMGM-7 and Pseudomonas sp. WH001 55 also produced melanin in the presence of L-tyrosine (Ganesh Kumar et al., 2013; Kiran et al., 2017). The prerequisite of L-tyrosine as the precursor is a common feature of eumelanin-synthesizing organisms (Tran-Ly et al., 2020). It was observed that beef extract was consumed more readily by the isolate and facilitated the high titer of melanin production (4.6 ± 0.1 g/l). A similar requirement of beef extract for enhanced melanin production (5.29 g/l) by S. lusitanus DMZ3 was reported previously (Madhusudhan et al., 2014). Copper ions are important cofactors for the functioning of melanogenic enzymes such as tyrosinases and laccases (Sendovski et al., 2011; Yang et al., 2017). The addition of CuCl2 (0.01%) significantly increased the yield of melanin produced by the isolate P. otitidis DDB2 by 0.7 g/l. Enhanced production of melanin can also be achieved with other metals by inducing stress in microorganisms (Gowri and Srivastava, 1996). For instance, Auricularia auricula and Streptomyces glaucescens NEAE-H require magnesium and iron salts, respectively, for enhanced melanin production (Sun et al., 2016; El Naggar and El-Ewasy, 2017). Surwase et al. (2013) reported improved production of melanin (6.8 g/l) by Brevundimonas sp. SGJ in the presence of L-tyrosine, tryptone, and CuSO4 after 54 h of incubation. In another study, the fungus Armillaria cepistipes produced the maximum amount of melanin (27.98 g/l) after 161 days of incubation and in the presence of carbon-nitrogen sources such as glucose and yeast extract along with metal salts of copper, iron, and magnesium (Ribera et al., 2019). The present study showed the maximum production of melanin (5.3 ± 0.1 g/l) by P. otitidis DDB2 with minimum medium ingredients and a short period of incubation (24 h) as compared to those reported earlier.
The extracted and purified melanin from P. otitidis DDB2 was found to be insoluble in most of the solvents tested, and this property of poor solubility is peculiar to melanin. The precipitation of melanin after reacting with FeCl3 indicated the presence of polyphenolic groups (Pralea et al., 2019). The melanin obtained in this study was degraded by alkaline H2O2, which was indicated by its bleaching. Purified melanin showed higher absorbance in UV range (Fig. 4), which decreased gradually with further increase in wavelength toward the visible region; this attribute is unique to melanin (Meredith and Sarna, 2006; Al Khatib et al., 2018). We characterized melanin extracted from P. otitidis DDB2 by other methods such as thermal stability, elemental analysis, FT-IR, and NMR as reported earlier (Seelam et al., 2021). Our previous studies on the characterization of melanin revealed its basic constituent as 5,6-dihydroxy indole, 2-carboxylic acid (Seelam et al., 2021). The three peaks observed in chromatogram of HPLC may be attributed to the oligomers of melanin with varying number of monomeric units. The broad spot developed on chromatogram of TLC and multiple peaks observed in HPLC show the heterogenous nature of melanin under study. It is a common feature of natural melanin to exhibit the property of heterogeneity and insolubility due to the complex arrangement of its molecular structure (Pralea et al., 2019).
In the present study, no antibacterial activity of melanin was detected against the tested pathogens E. coli, P. aeruginosa, S. entericus, and B. subtilis. The diameters of inhibition were very low as compared to the positive control streptomycin. Antibacterial activity of melanin in the present study showed contrasting results to earlier reports. For instance, melanin obtained from the fungus Schizophyllum communae exhibited antibacterial activity against E. coli, Klebsiella pneumoniae, Proteus sp., and Pseudomonas fluorescens at the concentration range of 50–200 μg (Arun et al., 2015). In another study, melanin (30 μg/disc) from Streptomyces sp. showed antibacterial activity against P. aeruginosa and Vibrio parahaemolyticus (Sivaperumal et al., 2014). Similarly, purified melanin from Actinoalloteichu s sp. MA-32 exhibited antibacterial activity against B. subtilis and E. coli at the concentration of 30 and 50 μg/disc (Manivasagan et al., 2013). Lopusiewicz (2018) reported the antibacterial activity of purified melanin extracted from Armillaria sp. against B. cereus, B. subtilis, Enterococcus faecalis, and P. aeruginosa. However, no activity was observed against B. anthrophaeus, E. coli, Micrococcus luteus, P. putida, and S. aureus. These incongruities in the antibacterial activity of melanin may result from the differences in the composition and molecular structure of melanin (Correa et al., 2017).
Recent advances in gene sequencing approaches and the development of computational biology tools have enabled the researchers to study the diversity of genes and understand the evolutionary relationship among different species. In the present study, an attempt was made to comparatively analyze tyrosinase gene and protein sequences of selected organisms with H. sapiens. The rationale underlying the selection of organisms for this study was that the potential organism in the present study was identified as Pseudomonas sp., and hence, different species of the same genus were selected for comparative analysis. Several studies have shown that the other common microbial sources for tyrosinase or melanin are Streptomyces (Madhusudhan et al., 2014; El Naggar and El-Ewasy 2017), Vibrio (Vijayan et al., 2017; Kurian and Bhat, 2018), or fungal species (Mattoon et al., 2021), and hence, they were also included in the study. S. officinalis was selected because this is the only source of commercially available natural melanin (Guo et al., 2014). The TYRP1 sequences of H. sapiens involved in melanin synthesis, maintenance of melanosome structure, and stabilization of tyrosinase were also selected as parameters to determine how likely TYRP1 and tyrosinase sequences of H. sapiens are related despite their location on different chromosomes (Zheng et al., 2010). However, the sequences of H. sapiens used as the out group were arbitrarily selected.
The distance matrix of the aligned sequences showed that the TYRP1 gene of H. sapiens (34.7%) is closer to tyrosinase of H. sapiens, followed by S. officinalis (21.9%) and T. marneffei ATCC18224 (20.5%). The protein sequences of TYRP1 of H. sapiens showed 39.3% identity, followed by S. azureus with 22.9% identity and S. clavuligerus with 22.7% identity with the tyrosinase protein of H. sapiens. The central domains of most of the species in the present study showed 100% identity with that of CCD of H. sapiens. Although a large variation was observed in the tyrosinase gene sequences of selected organisms, a 100% similarity in their central domains strongly implicate that the functional domains of the tyrosinase protein are highly conserved irrespective of their taxonomic positions. In silico studies provided insights on the variability of sequences and applicability of melanin to closely related species. Researchers from China have performed a similar analysis of TYRP1 among different species and showed that there were two evolutionarily conserved noncoding regions present upstream to the initiation sites of translation. They also reported the amino acids mutated within the species and the amino acids highly conserved among the species (Zheng et al., 2010). Therefore, the use of bioinformatics tools in scientific research aid in better understanding of evolutionary changes in a particular gene sequence and its directionality.
Conclusions
Melanin has recently gained significance for application in green technology and material science as an additive that can substantially enhance the performance of other materials. In light of its increasing demand, in the present study, we report the screening, isolation, and production of melanin by a potential strain of P. otitidis DDB2. The isolated melanin belonged to the eumelanin class with the maximum absorption range in the ultraviolet region, and hence, it could be used as a natural pigment in sunscreen products. Furthermore, in silico studies of melanogenic gene sequences provided us insights into the applicability of melanin to humans due to the evolutionary relatedness between the tyrosinase genes of humans and the selected Pseudomonas sp.